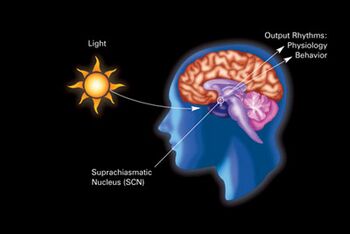
Biology:Suprachiasmatic nucleus
| Suprachiasmatic nucleus | |
|---|---|
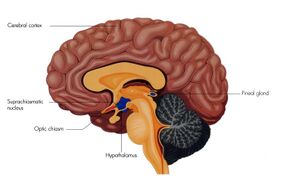 Suprachiasmatic nucleus in green | |
| Details | |
| Identifiers | |
| Latin | nucleus suprachiasmaticus |
| Anatomical terms of neuroanatomy | |
The suprachiasmatic nucleus or nuclei (SCN) is a small region of the brain in the hypothalamus, situated directly above the optic chiasm. The SCN is the principal circadian pacemaker in mammals, responsible for generating circadian rhythms.[1][2] Reception of light inputs from photosensitive retinal ganglion cells allow the SCN to coordinate the subordinate cellular clocks of the body and entrain to the environment.[1][3] The neuronal and hormonal activities it generates regulate many different body functions in an approximately 24-hour cycle.
The idea that the SCN is the main circadian pacemaker in mammals was proposed by Robert Moore, who conducted experiments using radioactive amino acids to find where the termination of the retinohypothalamic projection occurs in rodents.[4][5] Early lesioning experiments in mouse, guinea pig, cat, and opossum established how removal of the SCN results in ablation of circadian rhythm in mammals.[4]
Moreover, the SCN interacts with many other regions of the brain. It contains several cell types and several different peptides (including vasopressin and vasoactive intestinal peptide) and neurotransmitters.
Disruptions or damage to the SCN has been associated with different mood disorders and sleep disorders, suggesting the significance of the SCN in regulating circadian timing[6]
Neuroanatomy
The SCN is situated in the anterior part of the hypothalamus immediately dorsal, or superior (hence supra) to the optic chiasm bilateral to (on either side of) the third ventricle. It consists of two nuclei composed of approximately 10,000 neurons.[7]
The morphology of the SCN is species dependent.[8] Distribution of different cell phenotypes across specific SCN regions, such as the concentration of VP-IR neurons, can cause the shape of the SCN to change.[8]
The nucleus can be divided into ventrolateral and dorsolateral portions, also known as the core and shell, respectively.[7] These regions differ in their expression of the clock genes, the core expresses them in response to stimuli whereas the shell expresses them constitutively.
In terms of projections, the core receives innervation via three main pathways, the retinohypothalamic tract, geniculohypothalamic tract, and projections from some raphe nuclei.[8] The dorsomedial SCN is mainly innervated by the core and also by other hypothalamic areas. Lastly, its output is mainly to the subparaventricular zone and dorsomedial hypothalamic nucleus which both mediate the influence SCN exerts over circadian regulation of the body.[8]
The most abundant peptides found within the SCN are arginine-vasopressin (AVP), vasoactive intestinal polypeptide (VIP), and peptide histidine-isoleucine (PHI). Each of these peptides are localized in different regions. Neurons with AVP are found dorsomedially, whereas VIP-containing and PHI-containing neurons are found ventrolaterally.[9]
Circadian clock
Different organisms such as bacteria,[10] plants, fungi, and animals, show genetically based near-24-hour rhythms. Although all of these clocks appear to be based on a similar type of genetic feedback loop, the specific genes involved are thought to have evolved independently in each kingdom. Many aspects of mammalian behavior and physiology show circadian rhythmicity, including sleep, physical activity, alertness, hormone levels, body temperature, immune function, and digestive activity. Early experiments on the function of the SCN involved lesioning the SCN in hamsters.[11] SCN lesioned hamsters lost their daily activity rhythms.[11] Further, when the SCN of a hamster was transplanted into an SCN lesioned hamster, the hamster adopted the rhythms of the hamster from which the SCN was transplanted.[11] Together, these experiments suggest that the SCN is sufficient for generating circadian rhythms in hamsters.
Later studies have shown that skeletal, muscle, liver, and lung tissues in rats generate 24-hour rhythms, which dampen over time when isolated in a dish, where the SCN maintains its rhythms.[12] Together, these data suggest a model whereby the SCN maintains control across the body by synchronizing "slave oscillators," which exhibit their own near-24-hour rhythms and control circadian phenomena in local tissue.[13]
The SCN receives input from specialized photosensitive ganglion cells in the retina via the retinohypothalamic tract.[14] Neurons in the ventrolateral SCN (vlSCN) have the ability for light-induced gene expression. Melanopsin-containing ganglion cells in the retina have a direct connection to the ventrolateral SCN via the retinohypothalamic tract.[14] When the retina receives light, the vlSCN relays this information throughout the SCN allowing entrainment, synchronization, of the person's or animal's daily rhythms to the 24-hour cycle in nature.[14] The importance of entraining organisms, including humans, to exogenous cues such as the light/dark cycle, is reflected by several circadian rhythm sleep disorders, where this process does not function normally.[15]
Neurons in the dorsomedial SCN (dmSCN) are believed to have an endogenous 24-hour rhythm that can persist under constant darkness (in humans averaging about 24 hours 11 min).[16] A GABAergic mechanism is involved in the coupling of the ventral and dorsal regions of the SCN.[17]
Circadian rhythms of endothermic (warm-blooded) and ectothermic (cold-blooded) vertebrates
Information about the direct neuronal regulation of metabolic processes and circadian rhythm-controlled behaviors is not well known among either endothermic or ectothermic vertebrates, although extensive research has been done on the SCN in model animals such as the mammalian mouse and ectothermic reptiles, particularly lizards. The SCN is known to be involved not only in photoreception through innervation from the retinohypothalamic tract, but also in thermoregulation of vertebrates capable of homeothermy as well as regulating locomotion and other behavioral outputs of the circadian clock within ectothermic vertebrates.[18] The behavioral differences between both classes of vertebrates when compared to the respective structures and properties of the SCN as well as various other nuclei proximate to the hypothalamus provide insight into how these behaviors are the consequence of differing circadian regulation. Ultimately, many neuroethological studies must be done to completely ascertain the direct and indirect roles of the SCN on circadian-regulated behaviors of vertebrates.
The SCN of endotherms and ectotherms
In general, external temperature does not influence endothermic animal circadian rhythm because of the ability of these animals to keep their internal body temperature constant through homeostatic thermoregulation; however, peripheral oscillators (see Circadian rhythm) in mammals are sensitive to temperature pulses and will experience resetting of the circadian clock phase and associated genetic expression, suggesting how peripheral circadian oscillators may be separate entities from one another despite having a master oscillator within the SCN.[18] Furthermore, when individual neurons of the SCN from a mouse were treated with heat pulses, a similar resetting of oscillators was observed, but when an intact SCN was treated with the same heat pulse treatment the SCN was resistant to temperature change by exhibiting an unaltered circadian oscillating phase.[18] In ectothermic animals, particularly the ruin lizard, Podarcis siculus, temperature has been shown to affect the circadian oscillators within the SCN.[19] This reflects a potential evolutionary relationship among endothermic and ectothermic vertebrates as ectotherms rely on environmental temperature to affect their circadian rhythms and behavior while endotherms have an evolved SCN that is resistant to external temperature fluctuations and uses photoreception as a means for entraining the circadian oscillators within their SCN.[18] In addition, the differences of the SCN between endothermic and ectothermic vertebrates suggest that the neuronal organization of the temperature-resistant SCN in endotherms is responsible for driving thermoregulatory behaviors in those animals differently from those of ectotherms, since they rely on external temperature for engaging in certain behaviors.
Behaviors controlled by the SCN of vertebrates
Significant research has been conducted on the genes responsible for controlling circadian rhythm, in particular within the SCN. Knowledge of the gene expression of Clock (Clk) and Period2 (Per2), two of the many genes responsible for regulating circadian rhythm within the individual cells of the SCN, has allowed for a greater understanding of how genetic expression influences the regulation of circadian rhythm-controlled behaviors.[20] Studies on thermoregulation of ruin lizards and mice have informed some connections between the neural and genetic components of both vertebrates when experiencing induced hypothermic conditions.[19] Certain findings have reflected how evolution of SCN both structurally and genetically has resulted in the engagement of characteristic and stereotyped thermoregulatory behavior in both classes of vertebrates.
- Mice: Among vertebrates, it is known that mammals are endotherms that are capable of homeostatic thermoregulation. It has been shown that mice display thermosensitivity within the SCN. However, the regulation of body temperature in hypothermic mice is more sensitive to the amount of light in their environment.[21] Even while fasted, mice in darkened conditions and experiencing hypothermia maintained a stable internal body temperature.[21] In light conditions, mice showed a drop in body temperature under the same fasting and hypothermic conditions. Through analyzing genetic expression of Clock genes in wild-type and knockout strains, as well as analyzing the activity of neurons within the SCN and connections to proximate nuclei of the hypothalamus in the aforementioned conditions, it has been shown that the SCN is the center of control for circadian body temperature rhythm.[21] This circadian control, thus, includes both direct and indirect influence of many of the thermoregulatory behaviors that mammals engage in to maintain homeostasis.
- Ruin lizards: Several studies have been conducted on the genes expressed in circadian oscillating cells of the SCN during various light and dark conditions, as well as effects from inducing mild hypothermia in reptiles. In terms of structure, the SCNs of lizards have a closer resemblance to those of mice, possessing a dorsomedial portion and a ventrolateral core.[22] However, genetic expression of the circadian-related Per2 gene in lizards is similar to that in reptiles and birds, despite the fact that birds have been known to have a distinct SCN structure consisting of a lateral and medial portion.[23] Studying the lizard SCN because of the lizard's small body size and ectothermy is invaluable to understanding how this class of vertebrates modifies its behavior within the dynamics of circadian rhythm, but it has not yet been determined whether the systems of cold-blooded vertebrates were slowed as a result of decreased activity in the SCN or showed decreases in metabolic activity as a result of hypothermia.[19]
Other signals from the retina
The SCN is one of many nuclei that receive nerve signals directly from the retina.
Some of the others are the lateral geniculate nucleus (LGN), the superior colliculus, the basal optic system, and the pretectum:
- The LGN passes information about color, contrast, shape, and movement on to the visual cortex and itself signals to the SCN.
- The superior colliculus controls the movement and orientation of the eye.
- The basal optic system also controls eye movements.[24]
- The pretectum controls the size of the pupil.
Genetic Basis of SCN Function
The SCN is the central circadian pacemaker of mammals, serving as the coordinator of mammalian circadian rhythms. Neurons in an intact SCN show coordinated circadian rhythms in electrical activity.[25] Neurons isolated from the SCN have been shown to produce and sustain circadian rhythms in vitro,[26] suggesting that each individual neuron of the SCN can function as an independent circadian oscillator at the cellular level.[27] Each cell of the SCN synchronizes its oscillations to the cells around it, resulting in a network of mutually reinforced and precise oscillations constituting the SCN master clock.[28]
Mammals
The SCN functions as a circadian biological clock in vertebrates including teleosts, reptiles, birds, and mammals.[29] In mammals, the rhythms produced by the SCN are driven by a transcription-translation negative feedback loop (TTFL) composed of interacting positive and negative transcriptional feedback loops.[30][31][32] Within the nucleus of an SCN cell, the genes Clock and Bmal1 (mop3) encode the BHLH-PAS transcription factors CLOCK and BMAL1 (MOP3), respectively. CLOCK and BMAL1 are positive activators that form CLOCK-BMAL1 heterodimers. These heterodimers then bind to E-boxes upstream of multiple genes, including per and cry, to enhance and promote their transcription and eventual translation.[20][32] In mammals, there are three known homologs for the period gene in Drosophila, namely per1, per2, and per3.
As per and cry are transcribed and translated into PER and CRY, the proteins accumulate and form heterodimers in the cytoplasm. The heterodimers are phosphorylated at a rate that determines the length of the transcription-translation feedback loop (TTFL) and then translocate back into the nucleus where the phosphorylated PER-CRY heterodimers act on CLOCK and/or BMAL1 to inhibit their activity. Although the role of phosphorylation in the TTFL mechanism is known, the specific kinetics are yet to be elucidated.[33] As a result, PER and CRY function as negative repressors and inhibit the transcription of per and cry. Over time, the PER-CRY heterodimers degrade and the cycle begins again with a period of about 24.5 hours.[34][35][36][32][37] The integral genes involved, termed “clock genes," are highly conserved throughout both SCN-bearing vertebrates like mice, rats, and birds as well as in non-SCN bearing animals such as Drosophila.[38]
Electrophysiology
Neurons in the SCN fire action potentials in a 24-hour rhythm, even under constant conditions.[39] At mid-day, the firing rate reaches a maximum, and, during the night, it falls again. Rhythmic expression of circadian regulatory genes in the SCN requires depolarization in the SCN neurons via calcium and cAMP.[39] Thus, depolarization of SCN neurons via cAMP and calcium contributes to the magnitude of the rhythmic gene expression in the SCN.[39]
Further, the SCN synchronizes nerve impulses which spread to various parasympathetic and sympathetic nuclei.[40] The sympathetic nuclei drive glucocorticoid output from the adrenal gland which activates Per1 in the body cells, thus resetting the circadian cycle of cells in the body.[40] Without the SCN, rhythms in body cells dampen over time, which may be due to lack of synchrony between cells.[39]
Many SCN neurons are sensitive to light stimulation via the retina.[41] The photic response is likely linked to effects of light on circadian rhythms. In addition, application of melatonin in live rats and isolated SCN cells can decrease the firing rate of these neurons.[42][43] Variances in light input due to jet lag, seasonal changes, and constant light conditions all change the firing rhythm in SCN neurons demonstrating the relationship between light and SCN neuronal functioning.[39]
Clinical significance
Irregular sleep-wake rhythm disorder
Irregular sleep-wake rhythm (ISWR) disorder is thought to be caused by structural damage to the SCN, decreased responsiveness of the circadian clock to light and other stimuli, and decreased exposure to light.[6][44] People who tend to stay indoors and limit their exposure to light experience decreased nocturnal melatonin production. The decrease in melatonin production at night corresponds with greater expression of SCN-generated wakefulness during night, causing irregular sleep patterns.[6]
Major depressive disorder
Major depressive disorder (MDD) has been associated with altered circadian rhythms.[45] Patients with MDD have weaker rhythms that express clock genes in the brain. When SCN rhythms were disturbed, anxiety-like behavior, weight gain, helplessness, and despair were reported in a study conducted with mice. Abnormal glucocorticoid levels occurred in mice with no Bmal1 expression in the SCN.[45]
Alzheimer's disease
The functional disruption of the SCN can be observed in early stages of Alzheimer's disease (AD).[46] Changes in the SCN and melatonin secretion are major factors that cause circadian rhythm disturbances. These disturbances cause the normal physiology of sleep to change, such as the biological clock and body temperature during rest.[46] Patients with AD experience insomnia, hypersomnia, and other sleep disorders as a result of the degeneration of the SCN and changes in critical neurotransmitter concentrations.[46]
See also
- Chronobiology
- Photosensitive ganglion cell
- Sense of time
- Retinohypothalamic tract
- Shift work sleep disorder
- Non-24-hour sleep–wake disorder
References
- ↑ 1.0 1.1 Hastings, Michael H.; Maywood, Elizabeth S.; Brancaccio, Marco (August 2018). "Generation of circadian rhythms in the suprachiasmatic nucleus" (in en). Nature Reviews Neuroscience 19 (8): 453–469. doi:10.1038/s41583-018-0026-z. ISSN 1471-0048. PMID 29934559. https://www.nature.com/articles/s41583-018-0026-z.
- ↑ Hastings, MH; Maywood, ES; Brancaccio, M (11 March 2019). "The Mammalian Circadian Timing System and the Suprachiasmatic Nucleus as Its Pacemaker.". Biology 8 (1). doi:10.3390/biology8010013. PMID 30862123.
- ↑ Weaver, David R.; Emery, Patrick (2013-01-01), Squire, Larry R.; Berg, Darwin; Bloom, Floyd E. et al., eds., "Chapter 39 - Circadian Timekeeping" (in en), Fundamental Neuroscience (Fourth Edition) (San Diego: Academic Press): pp. 819–845, ISBN 978-0-12-385870-2, https://www.sciencedirect.com/science/article/pii/B9780123858702000391, retrieved 2023-04-25
- ↑ 4.0 4.1 Klein, David C.; Moore, Robert Y.; Reppert, Steven M. (1991) (in en). Suprachiasmatic Nucleus: The Mind's Clock. Oxford University Press. ISBN 978-0-19-506250-2. https://books.google.com/books?id=8fgwFsmTBwgC&dq=suprachiasmatic+nucleus&pg=PR16.
- ↑ Moore, Robert Y. (2013-01-01), Gillette, Martha U., ed., "Chapter One - The Suprachiasmatic Nucleus and the Circadian Timing System" (in en), Progress in Molecular Biology and Translational Science, Chronobiology: Biological Timing in Health and Disease (Academic Press) 119: 1–28, doi:10.1016/B978-0-12-396971-2.00001-4, PMID 23899592, https://www.sciencedirect.com/science/article/pii/B9780123969712000014, retrieved 2023-04-25
- ↑ 6.0 6.1 6.2 Ma, Melinda A.; Morrison, Elizabeth H. (2023), "Neuroanatomy, Nucleus Suprachiasmatic", StatPearls (Treasure Island (FL): StatPearls Publishing), PMID 31536270, http://www.ncbi.nlm.nih.gov/books/NBK546664/, retrieved 2023-04-25
- ↑ 7.0 7.1 Ma, Melinda A.; Morrison, Elizabeth H. (2023), "Neuroanatomy, Nucleus Suprachiasmatic", StatPearls (Treasure Island (FL): StatPearls Publishing), PMID 31536270, http://www.ncbi.nlm.nih.gov/books/NBK546664/, retrieved 2023-04-09
- ↑ 8.0 8.1 8.2 8.3 Morin, Lawrence P. (May 2013). "Neuroanatomy of the extended circadian rhythm system". Experimental Neurology 243: 4–20. doi:10.1016/j.expneurol.2012.06.026. ISSN 1090-2430. PMID 22766204.
- ↑ Reuss, Stefan (1996-08-01). "Components and connections of the circadian timing system in mammals" (in en). Cell and Tissue Research 285 (3): 353–378. doi:10.1007/s004410050652. ISSN 1432-0878. PMID 8772150. https://doi.org/10.1007/s004410050652.
- ↑ "Functioning and robustness of a bacterial circadian clock". Molecular Systems Biology 3 (1): 90. 2007. doi:10.1038/msb4100128. PMID 17353932.
- ↑ 11.0 11.1 11.2 Ralph, Martin R.; Foster, Russell G.; Davis, Fred C.; Menaker, Michael (1990-02-23). "Transplanted Suprachiasmatic Nucleus Determines Circadian Period". Science 247 (4945): 975–978. doi:10.1126/science.2305266. ISSN 0036-8075. PMID 2305266. http://dx.doi.org/10.1126/science.2305266.
- ↑ Yamazaki, Shin; Numano, Rika; Abe, Michikazu; Hida, Akiko; Takahashi, Ri-ichi; Ueda, Masatsugu; Block, Gene D.; Sakaki, Yoshiyuki et al. (2000-04-28). "Resetting Central and Peripheral Circadian Oscillators in Transgenic Rats". Science 288 (5466): 682–685. doi:10.1126/science.288.5466.682. ISSN 0036-8075. PMID 10784453. http://dx.doi.org/10.1126/science.288.5466.682.
- ↑ "Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus". PLOS Computational Biology 3 (4): e68. April 2007. doi:10.1371/journal.pcbi.0030068. PMID 17432930. Bibcode: 2007PLSCB...3...68B.
- ↑ 14.0 14.1 14.2 Miller, Joseph D.; Morin, Lawrence P.; Schwartz, William J.; Moore, Robert Y. (1996). "New Insights Into the Mammalian Circadian Clock". Sleep 19 (8): 641–667. doi:10.1093/sleep/19.8.641. ISSN 1550-9109. PMID 8958635.
- ↑ "Circadian rhythm sleep disorders". The Medical Clinics of North America 88 (3): 631–51, viii. May 2004. doi:10.1016/j.mcna.2004.01.010. PMID 15087208.
- ↑ "Human Biological Clock Set Back an Hour" (in en-US). 1999-07-15. https://news.harvard.edu/gazette/story/1999/07/human-biological-clock-set-back-an-hour/.
- ↑ Azzi, A; Evans, JA; Leise, T; Myung, J; Takumi, T; Davidson, AJ; Brown, SA (18 January 2017). "Network Dynamics Mediate Circadian Clock Plasticity.". Neuron 93 (2): 441–450. doi:10.1016/j.neuron.2016.12.022. PMID 28065650.
- ↑ 18.0 18.1 18.2 18.3 "Temperature as a universal resetting cue for mammalian circadian oscillators". Science 330 (6002): 379–85. October 2010. doi:10.1126/science.1195262. PMID 20947768. Bibcode: 2010Sci...330..379B.
- ↑ 19.0 19.1 19.2 "Circadian expression of the clock gene Per2 is altered in the ruin lizard (Podarcis sicula) when temperature changes". Brain Research. Molecular Brain Research 133 (2): 281–5. February 2005. doi:10.1016/j.molbrainres.2004.10.014. PMID 15710245. http://doc.rero.ch/record/4325/files/1_albrecht_cec.pdf.
- ↑ 20.0 20.1 Gekakis, N.; Staknis, D.; Nguyen, H. B.; Davis, F. C.; Wilsbacher, L. D.; King, D. P.; Takahashi, J. S.; Weitz, C. J. (1998-06-05). "Role of the CLOCK protein in the mammalian circadian mechanism". Science 280 (5369): 1564–1569. doi:10.1126/science.280.5369.1564. ISSN 0036-8075. PMID 9616112. Bibcode: 1998Sci...280.1564G. https://pubmed.ncbi.nlm.nih.gov/9616112/.
- ↑ 21.0 21.1 21.2 "Thermoregulation in the cold changes depending on the time of day and feeding condition: physiological and anatomical analyses of involved circadian mechanisms". Neuroscience 164 (3): 1377–86. December 2009. doi:10.1016/j.neuroscience.2009.08.040. PMID 19703527.
- ↑ "Pattern of organization of primary visual pathways in the European lizard Podarcis sicula Rafinesque". Journal für Hirnforschung 34 (3): 361–74. 1993. PMID 7505790.
- ↑ "Spatial and temporal variation of passer Per2 gene expression in two distinct cell groups of the suprachiasmatic hypothalamus in the house sparrow (Passer domesticus)". The European Journal of Neuroscience 16 (3): 429–36. August 2002. doi:10.1046/j.1460-9568.2002.02102.x. PMID 12193185.
- ↑ "The accessory optic system: basic organization with an update on connectivity, neurochemistry, and function". Neuroanatomy of the Oculomotor System. Progress in Brain Research. 151. 2006. pp. 407–40. doi:10.1016/S0079-6123(05)51013-6. ISBN 9780444516961. https://escholarship.org/content/qt3v25z604/qt3v25z604.pdf?t=lnpyco.
- ↑ Prosser, R. A. (February 1998). "In vitro circadian rhythms of the mammalian suprachiasmatic nuclei: comparison of multi-unit and single-unit neuronal activity recordings". Journal of Biological Rhythms 13 (1): 30–38. doi:10.1177/074873098128999899. ISSN 0748-7304. PMID 9486841. https://pubmed.ncbi.nlm.nih.gov/9486841/.
- ↑ Herzog, E. D.; Takahashi, J. S.; Block, G. D. (December 1998). "Clock controls circadian period in isolated suprachiasmatic nucleus neurons". Nature Neuroscience 1 (8): 708–713. doi:10.1038/3708. ISSN 1097-6256. PMID 10196587. https://pubmed.ncbi.nlm.nih.gov/10196587/.
- ↑ Honma, Sato; Ono, Daisuke; Suzuki, Yohko; Inagaki, Natsuko; Yoshikawa, Tomoko; Nakamura, Wataru; Honma, Ken-Ichi (2012). Suprachiasmatic nucleus: cellular clocks and networks. Progress in Brain Research. 199. pp. 129–141. doi:10.1016/B978-0-444-59427-3.00029-0. https://pubmed.ncbi.nlm.nih.gov/22877663/#:~:text=The%20suprachiasmatic%20nucleus%20(SCN),,as%20in%20an%20organotypic%20slice..
- ↑ Welsh, David K.; Takahashi, Joseph S.; Kay, Steve A. (2010). "Suprachiasmatic nucleus: cell autonomy and network properties". Annual Review of Physiology 72: 551–577. doi:10.1146/annurev-physiol-021909-135919. ISSN 1545-1585. PMID 20148688.
- ↑ Patton, Andrew P.; Hastings, Michael H. (2018-08-06). "The suprachiasmatic nucleus". Current Biology 28 (15): R816–R822. doi:10.1016/j.cub.2018.06.052. ISSN 1879-0445. PMID 30086310.
- ↑ Buhr, Ethan D.; Takahashi, Joseph S. (2013). "Molecular Components of the Mammalian Circadian Clock". Circadian Clocks. Handbook of Experimental Pharmacology. 217. pp. 3–27. doi:10.1007/978-3-642-25950-0_1. ISBN 978-3-642-25949-4.
- ↑ Shearman, Lauren P.; Sriram, Sathyanarayanan; Weaver, David R.; Maywood, Elizabeth S.; Chaves, Inẽs; Zheng, Binhai; Kume, Kazuhiko; Lee, Cheng Chi et al. (2000). "Interacting Molecular Loops in the Mammalian Circadian Clock". Science 288 (5468): 1013–1019. doi:10.1126/science.288.5468.1013. ISSN 0036-8075. PMID 10807566. Bibcode: 2000Sci...288.1013S. https://www.academia.edu/15431872.
- ↑ 32.0 32.1 32.2 Reppert, Steven M.; Weaver, David R. (2002-08-29). "Coordination of circadian timing in mammals". Nature 418 (6901): 935–941. doi:10.1038/nature00965. ISSN 0028-0836. PMID 12198538. Bibcode: 2002Natur.418..935R. https://pubmed.ncbi.nlm.nih.gov/12198538/.
- ↑ Herzog, Erik D.; Hermanstyne, Tracey; Smyllie, Nicola J.; Hastings, Michael H. (2017-01-03). "Regulating the Suprachiasmatic Nucleus (SCN) Circadian Clockwork: Interplay between Cell-Autonomous and Circuit-Level Mechanisms". Cold Spring Harbor Perspectives in Biology 9 (1): a027706. doi:10.1101/cshperspect.a027706. ISSN 1943-0264. PMID 28049647.
- ↑ Kume, K.; Zylka, M. J.; Sriram, S.; Shearman, L. P.; Weaver, D. R.; Jin, X.; Maywood, E. S.; Hastings, M. H. et al. (1999-07-23). "mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop". Cell 98 (2): 193–205. doi:10.1016/s0092-8674(00)81014-4. ISSN 0092-8674. PMID 10428031.
- ↑ Okamura, H.; Miyake, S.; Sumi, Y.; Yamaguchi, S.; Yasui, A.; Muijtjens, M.; Hoeijmakers, J. H.; van der Horst, G. T. (1999-12-24). "Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock". Science 286 (5449): 2531–2534. doi:10.1126/science.286.5449.2531. ISSN 0036-8075. PMID 10617474. https://pubmed.ncbi.nlm.nih.gov/10617474/.
- ↑ Gao, Peng; Yoo, Seung-Hee; Lee, Kyung-Jong; Rosensweig, Clark; Takahashi, Joseph S.; Chen, Benjamin P.; Green, Carla B. (2013-12-06). "Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length". The Journal of Biological Chemistry 288 (49): 35277–35286. doi:10.1074/jbc.M113.509604. ISSN 1083-351X. PMID 24158435.
- ↑ Matsumura, Ritsuko; Tsuchiya, Yoshiki; Tokuda, Isao; Matsuo, Takahiro; Sato, Miho; Node, Koichi; Nishida, Eisuke; Akashi, Makoto (2014-11-14). "The mammalian circadian clock protein period counteracts cryptochrome in phosphorylation dynamics of circadian locomotor output cycles kaput (CLOCK)". The Journal of Biological Chemistry 289 (46): 32064–32072. doi:10.1074/jbc.M114.578278. ISSN 1083-351X. PMID 25271155.
- ↑ Cassone, Vincent M. (January 2014). "Avian circadian organization: a chorus of clocks". Frontiers in Neuroendocrinology 35 (1): 76–88. doi:10.1016/j.yfrne.2013.10.002. ISSN 1095-6808. PMID 24157655.
- ↑ 39.0 39.1 39.2 39.3 39.4 Welsh, David K.; Takahashi, Joseph S.; Kay, Steve A. (2010-03-17). "Suprachiasmatic Nucleus: Cell Autonomy and Network Properties" (in en). Annual Review of Physiology 72 (1): 551–577. doi:10.1146/annurev-physiol-021909-135919. ISSN 0066-4278. PMID 20148688.
- ↑ 40.0 40.1 Okamura, H. (2007). "Suprachiasmatic Nucleus Clock Time in the Mammalian Circadian System" (in en). Cold Spring Harbor Symposia on Quantitative Biology 72 (1): 551–556. doi:10.1101/sqb.2007.72.033. ISSN 0091-7451. PMID 18419314.
- ↑ Morin, L.P.; Allen, C.N. (2006). "The circadian visual system, 2005" (in en). Brain Research Reviews 51 (1): 1–60. doi:10.1016/j.brainresrev.2005.08.003. PMID 16337005. https://linkinghub.elsevier.com/retrieve/pii/S0165017305001165.
- ↑ van den Top, M; Buijs, R.M; Ruijter, J.M; Delagrange, P; Spanswick, D; Hermes, M.L.H.J (2001). "Melatonin generates an outward potassium current in rat suprachiasmatic nucleus neurones in vitro independent of their circadian rhythm" (in en). Neuroscience 107 (1): 99–108. doi:10.1016/S0306-4522(01)00346-3. PMID 11744250. https://linkinghub.elsevier.com/retrieve/pii/S0306452201003463.
- ↑ Yang, Jing; Jin, Hui Juan; Mocaër, Elisabeth; Seguin, Laure; Zhao, Hua; Rusak, Benjamin (2016-06-15). "Agomelatine affects rat suprachiasmatic nucleus neurons via melatonin and serotonin receptors" (in en). Life Sciences 155: 147–154. doi:10.1016/j.lfs.2016.04.035. ISSN 0024-3205. PMID 27269050. https://www.sciencedirect.com/science/article/pii/S0024320516302557.
- ↑ Zee, Phyllis C.; Vitiello, Michael V. (2009-06-01). "Circadian Rhythm Sleep Disorder: Irregular Sleep Wake Rhythm" (in en). Sleep Medicine Clinics. Basics of Circadian Biology and Circadian Rhythm Sleep Disorders 4 (2): 213–218. doi:10.1016/j.jsmc.2009.01.009. ISSN 1556-407X. PMID 20160950.
- ↑ 45.0 45.1 Landgraf, Dominic; Long, Jaimie E.; Proulx, Christophe D.; Barandas, Rita; Malinow, Roberto; Welsh, David K. (2016-12-01). "Genetic Disruption of Circadian Rhythms in the Suprachiasmatic Nucleus Causes Helplessness, Behavioral Despair, and Anxiety-like Behavior in Mice" (in en). Biological Psychiatry. Novel Signaling Mechanisms in Depression 80 (11): 827–835. doi:10.1016/j.biopsych.2016.03.1050. ISSN 0006-3223. PMID 27113500.
- ↑ 46.0 46.1 46.2 Weldemichael, Dawit A.; Grossberg, George T. (2010-09-02). "Circadian Rhythm Disturbances in Patients with Alzheimer's Disease: A Review" (in en). International Journal of Alzheimer's Disease 2010: e716453. doi:10.4061/2010/716453. ISSN 2090-8024. PMID 20862344.
External links
 |