Biology:KcsA potassium channel
KcsA (K channel of streptomyces A) is a prokaryotic potassium channel from the soil bacterium Streptomyces lividans that has been studied extensively in ion channel research. The pH[1] activated protein possesses two transmembrane segments and a highly selective pore region, responsible for the gating and shuttling of K+ ions out of the cell.[2] The amino acid sequence found in the selectivity filter of KcsA is highly conserved among both prokaryotic and eukaryotic K+ voltage channels;[1][3] as a result, research on KcsA has provided important structural and mechanistic insight on the molecular basis for K+ ion selection and conduction. As one of the most studied ion channels to this day, KcsA is a template for research on K+ channel function and its elucidated structure underlies computational modeling of channel dynamics for both prokaryotic and eukaryotic species.[4]
| KcsA Potassium Channel | |||||||||
|---|---|---|---|---|---|---|---|---|---|
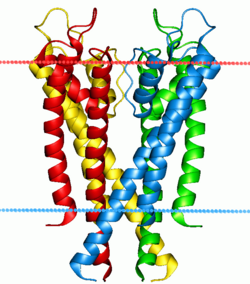 The four subunits forming the channel are drawn in different colors. They surround a central pore, guarded by the selectivity filter made up of the P-loops from each of the subunits. The blue and red dots indicate the boundaries of the lipid bilayer. | |||||||||
| Identifiers | |||||||||
| Symbol | KcsA | ||||||||
| Pfam | PF07885 | ||||||||
| InterPro | IPR013099 | ||||||||
| SCOP2 | 1bl8 / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 8 | ||||||||
| OPM protein | 1r3j | ||||||||
| |||||||||
History
KcsA was the first potassium ion channel to be characterized using x-ray crystallography by Roderick MacKinnon and his colleagues in 1998. In the years leading up to this, research on the structure of K+ channels was centered on the use of small toxin binding to reveal the location of the pore and selectivity filter among channel residues. MacKinnon's group theorized the tetrameric arrangement of the transmembrane segments, and even suggested presence of pore-forming “loops” in the filter region made of short segments of amino acids that interacted with K+ ions passing through the channel[5] The discovery of strong sequence homology between KcsA and other channels in the Kv family, including the Shaker protein, attracted the attention of the scientific community especially as the K+ channel signature sequence began to appear in other prokaryotic genes. The simplicity of the two transmembrane helices in KcsA, as opposed to the six in many eukaryotic ion channels, also provided a method to understand the mechanisms of K+ channels conduction at a more rudimentary level, thereby providing even great impetus for the study of KcsA.
The crystal structure of KcsA was solved by the MacKinnon group in 1998 after discovery that removal of the C-terminus cytoplasmic domain of the native protein (residues 126–158) increases the stability of crystallized samples. A model of KcsA at the 3.2A resolution was produced that confirmed the tetrameric arrangement of the protein around a center pore, with one helix of each subunit facing the inside axis and the other facing outwards.[6] Three years later, a higher resolution model was produced by Morais-Cabral and Zhou after monoclonal Fab fragments were attached to KcsA crystals to further stabilize the channel.[7] In the early 2000s, evidence for the occupation of the selectivity filter by two K+ atom during the transport process emerged, based on energy and electrostatic calculations made to model the pore region. Continued investigation of the various opened and closed, inactive and active conformations of KcsA by other imaging methods such as ssNMR and EPR have since provided even more insight into channel structure and the forces gating the switch from channel inactivation to conduction.
In 2007, Riek et al. showed that the channel opening that results from titrating the ion channel from pH 7 to pH 4, corresponds to conformational changes in two regions: transition to the ion-exchanging state of the selectivity filter, and the opening of the arrangement of TM2 at the C-terminus.[8] This model explains the ability of KcsA to simultaneous select for K+ ions while also gating electrical conductance. In 2011, the crystal structure of full length KcsA was resolved to reveal that hindrance by the previously truncated residues permits only straightforward expansion of the intercellular ion passage region of the protein. This research provides a more detailed look into the motion of separate channel regions during ion conduction.[9] In the present day, KcsA studies are focused on using the prokaryotic channel as a model for the channel dynamics of larger eukaryotic K+ channels, including hERG.
Structure

The structure of KcsA is that of an inverted cone, with a central pore running down the center made up of two transmembrane helices (the outer-helix M1 and the inner-helix M2), which span the lipid bilayer. The channel itself is a tetramer composed of four identical, single-domain subunits (each with two α-helices) arranged so that one M2 helix faces the central pore, while the other M1 helix faces the lipid membrane. The inner helices are tilted by about 25° in relation to the lipid membrane and are slightly kinked, opening up to face the outside of the cell like a flower.[6] These two TM helices are linked by a reentrant loop, dispersed symmetrically around a common axis corresponding to the central pore. The pore region spans approximately 30 amino acid residues and can be divided into three parts: a selectivity filter near the extracellular side, a dilated water-filled cavity at the center, and a closed gate near the cytoplasmic side formed by four packed M2 helices.[6] This architecture is found to be highly conserved in the potassium channel family[10][11] in both eukaryotes and prokaryotes.
The overall length of the pore is 45 Å, and its diameter varies considerably within the distinct regions of the inner tunnel. Travelling from the intracellular region outwards (bottom to top in the picture) the pore begins with a gate region formed by M2 helices at 18 Å in diameter, and then opens into a wide cavity (~10 Å across) near the middle of the membrane.[6] In these regions, K+ ions are in contact with surrounding water molecules but when they enter the channel from the selectivity filter at the top, the cavity is so narrow that K+ ions must shed any hydrating waters in order to enter the cell.[6] In regards to the amino acid composition of the pore-lining residues within KcsA, the side chains lining the internal pore and cavity are predominantly hydrophobic, but within the selectivity filter polar amino acids are present that contact the dehydrated K+ ions.
Selectivity filter
The wider end of the cone corresponds to the extracellular mouth of the channel made up of pore helices, plus a selectivity filter that is formed by a TVGYG sequence, (Threonine, Valine, Glycine, Tyrosine, Glycine), characteristic of potassium channels.[12] Within this region, coordination between the TVGYG amino acids and incoming K+ ions allows for conduction of ions through the channel. The selectivity filter of KcsA contains four ion binding sites, although it is proposed that only two of these four positions are occupied at one time. The selectivity filter is about 3 Å in diameter.[13] though molecular dynamics simulations suggest the filter is flexible.[14] The presence of TVGYG in the filter region of KcsA is conserved even in more complex eukaryotic channels, thus making KcsA an optimal system for studying K+ channel conductance across species.
Function
The KcsA channel is considered a model channel because the KcsA structure provides a framework for understanding K+ channel conduction, which has three parts: Potassium selectivity, channel gating by pH sensitivity, and voltage-gated channel inactivation. K+ ion permeation occurs at the upper selectivity filter region of the pore, while pH gating rises from the protonation of transmembrane helices at the end of the pore. At low pH, the M2 helix is protonated, shifting the ion channel from closed to open conformation.[15] As ions flow through the channel, voltage gating mechanisms are thought to induce interactions between Glu71 and Asp80 in the selectivity filter, which destabilize the conductive conformation and facilitate entry into a long-lived nonconducting state that resembles the C-type–inactivation of voltage-dependent channels.[16]
In the nonconducting conformation of KcsA at pH 7, K+ is bound tightly to coordinating oxygens of the selectivity filter and the four TM2 helices converge near the cytoplasmic junction to block the passage of any potassium ions.[8] At pH 4 however, KcsA undergoes millisecond-timescale conformational exchanges filter permeating and nonpermeating states and between the open and closed conformations of the M2 helices.[8] While these distinct conformational changes occur in separate regions of the channel, the molecular behavior of each region is linked by both electrostatic interactions and allostery.[8] The dynamics of this exchange stereochemical configurations in the filter provides the physical basis for simultaneous K+ conductance and gating.
K+selectivity
The sequence TVGYG is especially important for maintaining the potassium specificity of KcsA. The glycines in this selectivity filter sequence have dihedral angles that allow carbonyl oxygen atoms in the protein backbone of the filter to point in one direction, toward the ions along the pore.[5] The glycines and threonine coordinate with the K+ ion, while the side-chains of valine and tyrosine are directed into the protein core to impose geometric constraint on the filter. As a result, the KcsA tetramer harbors four equal spaced K+ binding sites, with each side composed of a cage formed by eight oxygen atoms that sit on the vertices of a cube. The oxygen atoms that surround K+ ions in the filter are arranged like the water molecules that encircle hydrated K+ ions in the cavity of the channel; this suggests that oxygen coordination and binding sites in the selectivity filter are paying for the energetic cost of K+ dehydration.[5] Because the Na+ ion is too small for these K+-sized binding sites, dehydration energy is not compensated and thus, the filter selects against other extraneous ions.[5] Additionally, the KcsA channel is blocked by Cs+ ions and gating requires the presence of Mg2+ ions.[1]
pH Sensitivity
The pH-dependent conductance of KcsA indicates that the opening of the ion channel occurs when the protein is exposed to a more acidic environment. NMR studies performed by the Riek group show that pH sensitivity occurs in both the C-terminal TM2 region of the protein as well as with Tyr78 and Gly79 residues in the selectivity filter. There is evidence to suggest that the main pH sensor is in the cytoplasmic domain. Exchanging negatively charged amino acids for neutral ones made the KcsA channel insensitive to pH even though there were no amino-acid changes at the transmembrane region.[17][18] In addition, between the pH of 6 and 7, histidine is one of the few titratable side chains of histidines; they are absent in the transmembrane and extracellular segments of TM2 but present at KcsA's C-terminus. This highlights a possible mechanism for the slow opening of KcsA which is particularly pH sensitive, especially as the conformational propagation of channel opening signal from the C-terminus to the selectivity filter could be important in coordinating the structural changes needed for conductance along the entire pore.
NMR studies also suggest that a complex hydrogen bond network between Tyr78, Gly79, Glu71 and Asp80 exists in the KcsA filter region, and further acts as a pH-sensitive trigger for conductance. The mutation of key residues in the region, including E71A, results in a large energy cost of 4 kcal mol−1, equivalent to the loss of the hydrogen bond between Glu71 and Tyr78 and the water-mediated hydrogen bond between Glu71 and Asp80 in KcsA(E71A). These studies further highlight the role of pH gating in KcsA channel function.
Voltage Gating
In 2006, the Perozo group proposed a mechanistic explanation for the effects of voltage fields on KcsA gating. After adding a depolarizing current to the channel, the reorientation of Glu71 towards the intracellular pore occurs, thereby disrupting the Glu71-Asp80 carboxyl-carboxylate pair that initially stabilizes the selectivity filter. The collapse of the filter region prevents entry into or facilitate exit from the inactivated state.[16] Glu71, a key part of the selectivity filter signature sequence that is conserved among K+ ion channels, plays a pivotal role in gating as its ability to reorient itself in the direction of the transmembrane voltage field is able to provide an explanation for voltage gating events in KcsA. The orientation of amino acids in the filter region might play significant physiological role in modulating potassium fluxes in eukaryotes and prokaryotes under steady-state conditions.[16]
Research
Function
The precise mechanism of potassium channel selectivity continues to be studied and debated and multiple models are used to describe different aspects of the selectivity. Models explaining selectivity based on field strength concept developed by George Eisenman[19] based on Coulomb's law have been applied to KcsA.[14][20] An alternative explanation for the selectivity of KcsA is based on the close-fit model (also known as the snug-fit model) developed by Francisco Bezanilla and Armstrong.[21] The main chain carbonyl oxygen atoms that make up the selectivity filter are held at a precise position that allows them to substitute for water molecules in the hydrated shell of the potassium ion, but they are too far from a sodium ion. Further work has studied thermodynamic differences in ion binding,[22] topological considerations,[23][24] and the number of continuous ion binding sites.[25]
In addition, a major limitation of crystal structure study and simulations has yet to be discussed: the best resolved and most applied crystal structure of KcsA appears to be that of the ‘closed' form of the channel. This is reasonable as the closed state of the channel is favored at neutral pH, at which the crystal structure was solved by X-ray crystallography. However, the dynamic behavior of KcsA makes analysis of the channel difficult as a crystal structure inevitably provides a static, spatially and temporally averaged image of a channel. To bridge the gap between molecular structure and physiological behavior, an understanding of the atomic resolution dynamics of potassium channels is required.
Applications
Due to the high sequence similarity between the pore of KcsA and other eukaryotic K+ ion channel proteins, KcsA has provided important insight into the behavior of other important voltage conducting proteins such as the drosophilla-derived Shaker and the human hERG potassium channel. KcsA has been used in mutagenesis studies to model the interactions between hERG and various drug compounds. Such tests can screen for drug-hERG channel interactions that cause acquired long QT syndrome, are essential for determining the cardiac safety of new medications.[26] In addition, homology models based on the closed state KcsA crystal structure have been generated computationally to construct a multiple state representation of the hERG cardiac K+ channel. Such models reveal the flexibility of the hERG channel and can consistently predict the binding affinity of a set of diverse ion channel-interacting ligands. Analysis of the complex ligand-hERG structures can be used to guide the synthesis of drug analogs with reduced hERG liability, based on drug structure and docking potential.[27]
See also
References
- ↑ 1.0 1.1 1.2 "A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans". The EMBO Journal 14 (21): 5170–8. Nov 1995. doi:10.1002/j.1460-2075.1995.tb00201.x. PMID 7489706.
- ↑ "Exploring the open pore of the potassium channel from Streptomyces lividans". FEBS Letters 462 (3): 447–452. 1999. doi:10.1016/S0014-5793(99)01579-3. PMID 10622743.
- ↑ "Overview of molecular relationships in the voltage-gated ion channel superfamily". Pharmacological Reviews 57 (4): 387–95. Dec 2005. doi:10.1124/pr.57.4.13. PMID 16382097.
- ↑ "Ion conduction and selectivity in K(+) channels". Annual Review of Biophysics and Biomolecular Structure 34: 153–71. 2005. doi:10.1146/annurev.biophys.34.040204.144655. PMID 15869387.
- ↑ 5.0 5.1 5.2 5.3 Roderick MacKinnon. "Nobel Lecture: Potassium Channels and the Atomic Basis of Selective Ion Conduction". Nobelprize.org. Nobel Media AB. https://www.nobelprize.org/nobel_prizes/chemistry/laureates/2003/mackinnon-lecture.html.
- ↑ 6.0 6.1 6.2 6.3 6.4 "The structure of the potassium channel: molecular basis of K+ conduction and selectivity". Science 280 (5360): 69–77. Apr 1998. doi:10.1126/science.280.5360.69. PMID 9525859. Bibcode: 1998Sci...280...69D.
- ↑ "Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution". Nature 414 (6859): 43–8. Nov 2001. doi:10.1038/35102009. PMID 11689936. Bibcode: 2001Natur.414...43Z.
- ↑ 8.0 8.1 8.2 8.3 "Conformational dynamics of the KcsA potassium channel governs gating properties". Nature Structural & Molecular Biology 14 (11): 1089–95. Nov 2007. doi:10.1038/nsmb1311. PMID 17922011.
- ↑ "Mechanism of activation gating in the full-length KcsA K+ channel". Proceedings of the National Academy of Sciences of the United States of America 108 (29): 11896–9. Jul 2011. doi:10.1073/pnas.1105112108. PMID 21730186. Bibcode: 2011PNAS..10811896U.
- ↑ "Ion conduction pore is conserved among potassium channels". Nature 413 (6858): 809–13. Oct 2001. doi:10.1038/35101535. PMID 11677598. Bibcode: 2001Natur.413..809L.
- ↑ "Potassium channel structures". Nature Reviews. Neuroscience 3 (2): 115–21. Feb 2002. doi:10.1038/nrn727. PMID 11836519.
- ↑ "Ion channels: from idea to reality". Nature Medicine 5 (10): 1105–9. Oct 1999. doi:10.1038/13415. PMID 10502800.
- ↑ "Potassium channels in myelinated nerve. Selective permeability to small cations". The Journal of General Physiology 61 (6): 669–86. Jun 1973. doi:10.1085/jgp.61.6.669. PMID 4541077.
- ↑ 14.0 14.1 "Ion selectivity in potassium channels". Biophysical Chemistry 124 (3): 279–91. Dec 2006. doi:10.1016/j.bpc.2006.05.033. PMID 16843584.
- ↑ "Molecular mechanism of pH sensing in KcsA potassium channels". Proceedings of the National Academy of Sciences of the United States of America 105 (19): 6900–5. May 2008. doi:10.1073/pnas.0800873105. PMID 18443286. Bibcode: 2008PNAS..105.6900T.
- ↑ 16.0 16.1 16.2 "Molecular determinants of gating at the potassium-channel selectivity filter". Nature Structural & Molecular Biology 13 (4): 311–8. Apr 2006. doi:10.1038/nsmb1069. PMID 16532009.
- ↑ "Role of the KcsA channel cytoplasmic domain in pH-dependent gating". Biophysical Journal 101 (9): 2157–62. Nov 2011. doi:10.1016/j.bpj.2011.09.024. PMID 22067153. Bibcode: 2011BpJ...101.2157H.
- ↑ "GCN4 enhances the stability of the pore domain of potassium channel KcsA". The FEBS Journal 275 (24): 6228–36. Dec 2008. doi:10.1111/j.1742-4658.2008.06747.x. PMID 19016844.
- ↑ "Cation selective glass electrodes and their mode of operation". Biophysical Journal 2 (2 Pt 2): 259–323. Mar 1962. doi:10.1016/S0006-3495(62)86959-8. PMID 13889686. Bibcode: 1962BpJ.....2..259E.
- ↑ "Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands". Nature 431 (7010): 830–4. Oct 2004. doi:10.1038/nature02943. PMID 15483608. Bibcode: 2004Natur.431..830N.
- ↑ "Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons". The Journal of General Physiology 60 (5): 588–608. Nov 1972. doi:10.1085/jgp.60.5.588. PMID 4644327.
- ↑ "Tuning ion coordination architectures to enable selective partitioning". Biophysical Journal 93 (4): 1093–9. Aug 2007. doi:10.1529/biophysj.107.107482. PMID 17513348. Bibcode: 2007BpJ....93.1093V.
- ↑ "The predominant role of coordination number in potassium channel selectivity". Biophysical Journal 93 (8): 2635–43. Oct 2007. doi:10.1529/biophysj.107.108167. PMID 17573427. Bibcode: 2007BpJ....93.2635T.
- ↑ "Selectivity in K+ channels is due to topological control of the permeant ion's coordinated state". Proceedings of the National Academy of Sciences of the United States of America 104 (22): 9260–5. May 2007. doi:10.1073/pnas.0700554104. PMID 17519335. Bibcode: 2007PNAS..104.9260B.
- ↑ "Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites". Proceedings of the National Academy of Sciences of the United States of America 108 (2): 598–602. Jan 2011. doi:10.1073/pnas.1013636108. PMID 21187421. Bibcode: 2011PNAS..108..598D.
- ↑ "Predicting drug-hERG channel interactions that cause acquired long QT syndrome". Trends in Pharmacological Sciences 26 (3): 119–24. Mar 2005. doi:10.1016/j.tips.2005.01.003. PMID 15749156.
- ↑ "A two-state homology model of the hERG K+ channel: application to ligand binding". Bioorganic & Medicinal Chemistry Letters 15 (6): 1737–41. Mar 2005. doi:10.1016/j.bmcl.2005.01.008. PMID 15745831.
 |


