Biology:AP-1 transcription factor
| AP-1 Proteins (Fos, ATF, JDP) | |
|---|---|
| Identifiers | |
| Symbol | AP-1 |
| InterPro | IPR000837 |
| Transcription factor Jun | |
|---|---|
| Identifiers | |
| Symbol | Leuzip_Jun |
| InterPro | IPR002112 |
Activator protein 1 (AP-1) is a transcription factor that regulates gene expression in response to a variety of stimuli, including cytokines, growth factors, stress, and bacterial and viral infections.[1] AP-1 controls a number of cellular processes including differentiation, proliferation, and apoptosis.[2] The structure of AP-1 is a heterodimer composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families.
History
AP-1 was first discovered as a TPA-activated transcription factor that bound to a cis-regulatory element of the human metallothionein IIa (hMTIIa) promoter and SV40.[3] The AP-1 binding site was identified as the 12-O-Tetradecanoylphorbol-13-acetate (TPA) response element (TRE) with the consensus sequence 5’-TGA G/C TCA-3’.[4] The AP-1 subunit Jun was identified as a novel oncoprotein of avian sarcoma virus, and Fos-associated p39 protein was identified as the transcript of the cellular Jun gene. Fos was first isolated as the cellular homologue of two viral v-fos oncogenes, both of which induce osteosarcoma in mice and rats.[5] Since its discovery, AP-1 has been found to be associated with numerous regulatory and physiological processes, and new relationships are still investigated.
Structure
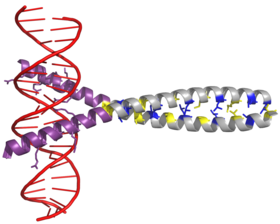
AP-1 transcription factor is assembled through the dimerization of a characteristic bZIP domain (basic region leucine zipper) in the Fos and Jun subunits. A typical bZIP domain consists of a “leucine zipper” region, and a “basic region”. The leucine zipper is responsible for dimerization of the Jun and Fos protein subunits. This structural motif twists two alpha helical protein domains into a “coiled coil,” characterized by a periodicity of 3.5 residues per turn and repetitive leucines appearing at every seventh position of the polypeptide chain. Due to the amino acid sequence and the periodicity of the helices, the leucine side chains are arranged along one face of the α helix and form a hydrophobic surface that modulates dimerization.[6] Hydrophobic residues additional to leucine also form the characteristic 3-4 repeat of α helices involved in “coiled-coil” interactions, and help contribute to the hydrophobic packing that drives dimerization. Together, this hydrophobic surface holds the two subunits together.[7][8]
The basic region of the bZIP domain is just upstream to the leucine zipper, and contains positively charged residues. This region interacts with DNA target sites.[9] Apart from the “leucine zipper” and the “basic region” which are important for dimerization and DNA-binding, the c-jun protein contains three short regions, which consist of clusters of negatively charged amino acids in its N-terminal half that are important for transcriptional activation in vivo.[10]
Dimerization happens between the products of the c-jun and c-fos protooncogenes, and is required for DNA-binding. Jun proteins can form both homo and heterodimers and therefore are capable of binding to DNA by themselves. However, Fos proteins do not dimerize with each other and therefore can only bind to DNA when bound with Jun.[11][12] The Jun-Fos heterodimer is more stable and has higher DNA-binding activity than Jun homodimers.
Function
AP-1 transcription factor has been shown to have a hand in a wide range of cellular processes, including cell growth, differentiation, and apoptosis. AP-1 activity is often regulated via post-translational modifications, DNA binding dimer composition, and interaction with various binding partners. AP-1 transcription factors are also associated with numerous physiological functions especially in determination of organisms’ life span and tissue regeneration. Below are some of the other important functions and biological roles AP-1 transcription factors have been shown to be involved in.
Cell growth, proliferation and senescence
The AP-1 transcription factor has been shown to play numerous roles in cell growth and proliferation. In particular, c-Fos and c-Jun seem to be major players in these processes. C-jun has been shown to be essential for fibroblast proliferation,[13] and levels of both AP-1 subunits have been shown to be expressed above basal levels during cell division.[14] C-fos has also been shown to increase in expression in response to the introduction of growth factors in the cell, further supporting its suggested involvement in the cell cycle. The growth factors TGF alpha, TGF beta, and IL2 have all been shown to stimulate c-Fos, and thereby stimulate cellular proliferation via AP-1 activation.[10]
Cellular senescence has been identified as "a dynamic and reversible process regulated by (in)activation of a predetermined enhancer landscape controlled by the pioneer transcription factor AP-1", which "defines the organizational principles of the transcription factor network that drives the transcriptional programme of senescent cells".[15][16]
Cellular differentiation
AP-1 transcription is deeply involved in the modulation of gene expression. Changes in cellular gene expression in the initiation of DNA synthesis and the formation of differentiated derivatives can lead to cellular differentiation.[10] AP-1 has been shown to be involved in cell differentiation in several systems. For example, by forming stable heterodimers with c-Jun, the bZIP region of c-Fos increases the binding of c-Jun to target genes whose activation is involved in the differentiation of chicken embryo fibroblasts (CEF).[17] It has also been shown to participate in endoderm specification.[18]
Apoptosis
AP-1 transcription factor is associated with a broad range of apoptosis related interactions. AP-1 activity is induced by numerous extracellular matrix and genotoxic agents, suggesting involvement in programmed cell death.[2] Many of these stimuli activate the c-Jun N-terminal kinases (JNKs) leading to the phosphorylation of Jun proteins and enhanced transcriptional activity of AP-1 dependent genes.[2] Increases in the levels of Jun and Fos proteins and JNK activity have been reported in scenarios in which cells undergo apoptosis. For example, inactivated c-Jun-ER cells show a normal morphology, while c-Jun-ER activated cells have been shown to be apoptotic.[19]
Regulation of AP-1
Increased AP-1 levels lead to increased transactivation of target gene expression. Regulation of AP-1 activity is therefore critical for cell function and occurs through specific interactions controlled by dimer-composition, transcriptional and post-translational events, and interaction with accessory proteins.[20]
AP-1 functions are heavily dependent on the specific Fos and Jun subunits contributing to AP-1 dimers.[10] The outcome of AP-1 activation is dependent on the complex combinatorial patterns of AP-1 component dimers.[2] The AP-1 complex binds to a palindromic DNA motif (5’-TGA G/C TCA-3’) to regulate gene expression, but specificity is dependent on the dimer composition of the bZIP subunit.[2]
Physiological relevance
AP-1 transcription factor has been shown to be involved in skin physiology, specifically in tissue regeneration. The process of skin metabolism is initiated by signals that trigger undifferentiated proliferative cells to undergo cell differentiation. Therefore, activity of AP-1 subunits in response to extracellular signals may be modified under conditions where the balance of keratinocyte proliferation and differentiation has to be rapidly and temporally altered.[21] The AP-1 transcription factor also has been shown to be involved in breast cancer cell growth through multiple mechanisms, including regulation of cyclin D1, E2F factors and their target genes. c-Jun, which is one of the AP-1 subunits, regulates the growth of breast cancer cells. Activated c-Jun is predominantly expressed at the invasive front in breast cancer and is associated with proliferation of breast cells.[22] Due to the AP-1 regulatory functions in cancer cells, AP-1 modulation is studied as a potential strategy for cancer prevention and therapy.[23][24][25]
Regulome
See also
- Activator protein
- Transcription factor
References
- ↑ "AP-1 subunits: quarrel and harmony among siblings". Journal of Cell Science 117 (Pt 25): 5965–73. December 2004. doi:10.1242/jcs.01589. PMID 15564374.
- ↑ 2.0 2.1 2.2 2.3 2.4 "A role for AP-1 in apoptosis: the case for and against". Biochimie 85 (8): 747–52. August 2003. doi:10.1016/j.biochi.2003.09.006. PMID 14585541.
- ↑ "Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40". Nature 325 (6102): 368–72. January 1987. doi:10.1038/325368a0. PMID 3027570. Bibcode: 1987Natur.325..368L.
- ↑ "Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor". Cell 49 (6): 729–39. June 1987. doi:10.1016/0092-8674(87)90611-8. PMID 3034432.
- ↑ "AP-1--Introductory remarks". Oncogene 20 (19): 2334–5. April 2001. doi:10.1038/sj.onc.1204416. PMID 11402330.
- ↑ "The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins". Science 240 (4860): 1759–64. June 1988. doi:10.1126/science.3289117. PMID 3289117. Bibcode: 1988Sci...240.1759L.
- ↑ "Evidence that the leucine zipper is a coiled coil". Science 243 (4890): 538–42. January 1989. doi:10.1126/science.2911757. PMID 2911757. Bibcode: 1989Sci...243..538O.
- ↑ "Preferential heterodimer formation by isolated leucine zippers from fos and jun". Science 245 (4918): 646–8. August 1989. doi:10.1126/science.2503872. PMID 2503872. Bibcode: 1989Sci...245..646O.
- ↑ "jun: oncogene and transcription factor". Advances in Cancer Research 55: 1–35. 1990. doi:10.1016/s0065-230x(08)60466-2. ISBN 9780120066551. PMID 2166997.
- ↑ 10.0 10.1 10.2 10.3 "The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 1072 (2–3): 129–57. December 1991. doi:10.1016/0304-419X(91)90011-9. PMID 1751545.
- ↑ "The role of the leucine zipper in the fos-jun interaction". Nature 336 (6200): 646–51. December 1988. doi:10.1038/336646a0. PMID 2974122. Bibcode: 1988Natur.336..646K.
- ↑ "DNA binding activities of three murine Jun proteins: stimulation by Fos". Cell 55 (5): 907–15. December 1988. doi:10.1016/0092-8674(88)90146-8. PMID 3142691.
- ↑ "AP-1 function and regulation". Current Opinion in Cell Biology 9 (2): 240–6. April 1997. doi:10.1016/S0955-0674(97)80068-3. PMID 9069263.
- ↑ "The Activating Protein-1 Transcriptional Complex: Essential and Multifaceted Roles in Bone". Clinical Reviews in Bone and Mineral Metabolism 4 (2): 107–122. 2006. doi:10.1385/BMM:4:2:107.
- ↑ "In and out from senescence". Nat Cell Biol 22 (7): 753–754. 2020. doi:10.1038/s41556-020-0540-x. PMID 32591745.
- ↑ "AP-1 imprints a reversible transcriptional programme of senescent cells". Nat Cell Biol 22 (7): 842–855. 2020. doi:10.1038/s41556-020-0529-5. PMID 32514071.
- ↑ "AP-1 as a regulator of cell life and death". Nature Cell Biology 4 (5): E131–6. May 2002. doi:10.1038/ncb0502-e131. PMID 11988758.
- ↑ "Epigenetic and transcriptional regulations prime cell fate before division during human pluripotent stem cell differentiation". Nature Communications 14 (405): 405. January 25, 2023. doi:10.1038/s41467-023-36116-9. PMID 36697417. PMC 9876972. Bibcode: 2023NatCo..14..405M. https://www.nature.com/articles/s41467-023-36116-9.pdf.
- ↑ "Induction of apoptosis by the transcription factor c-Jun". The EMBO Journal 16 (7): 1695–709. April 1997. doi:10.1093/emboj/16.7.1695. PMID 9130714.
- ↑ "Translational regulation mechanisms of AP-1 proteins". Mutation Research 682 (1): 7–12. July 2009. doi:10.1016/j.mrrev.2009.01.001. PMID 19167516.
- ↑ "Function and regulation of AP-1 subunits in skin physiology and pathology". Oncogene 20 (19): 2413–23. April 2001. doi:10.1038/sj.onc.1204380. PMID 11402337.
- ↑ "The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors". Oncogene 27 (3): 366–77. January 2008. doi:10.1038/sj.onc.1210643. PMID 17637753.
- ↑ "AP-1: a double-edged sword in tumorigenesis". Nature Reviews. Cancer 3 (11): 859–68. November 2003. doi:10.1038/nrc1209. PMID 14668816.
- ↑ "Targeting activator protein 1 signaling pathway by bioactive natural agents: Possible therapeutic strategy for cancer prevention and intervention". Pharmacological Research 128: 366–375. February 2018. doi:10.1016/j.phrs.2017.09.014. PMID 28951297.
- ↑ "Selective activator protein-1 inhibitor T-5224 prevents lymph node metastasis in an oral cancer model". Cancer Science 107 (5): 666–73. May 2016. doi:10.1111/cas.12914. PMID 26918517.
External links
 |



