Biology:NqrA-Marinomonas RNA motif
| nqrA-Marinomonas | |
|---|---|
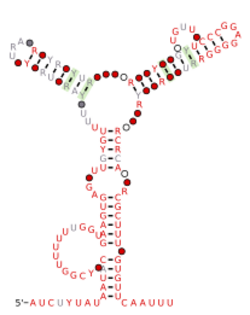 Consensus secondary structure and sequence conservation of nqrA-Marinomonas RNA | |
| Identifiers | |
| Symbol | nqrA-Marinomonas |
| Rfam | RF03035 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The nqrA-Marinomonas RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] nqrA-Marinomonas motif RNAs are found in Marinomonas.
All known nqrA-Marinomonas RNAs are found upstream of "nqr" operons whose genes encode subunits of the sodium-translocating NADH:quinone oxidoreductase, whose function is suspected to be to allow marine bacteria to form a sodium gradient. The name of the RNA motif is derived from the first gene in the operon, which is called nqrA. Based on their locations, it is reasonable to hypothesize that nqrA-Marinomonas RNAs function as cis-regulatory elements. However, promoter sequences occur upstream of the nqr operon in the distantly related organism Vibrio anguillarum.[2] The fact that these promoters do not occur upstream of (or in the same inter-genic region) as nqrA-Marinomonas RNAs raises questions about the putative cis-regulatory function of these RNAs.[1] However, it is possible that the promoters that were determined in Vibrio anguillarum are simply to diverged to be detected in Marinomonas species.
References
- ↑ 1.0 1.1 "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ "Transcriptional regulation of the Na+-NADH:quinone exidoreductase gene, nqr, in Vibrio anguillarum, a fish pathogene, in the stationary phase". Fisheries Science 73 (2): 378–355. 2007. doi:10.1111/j.1444-2906.2007.01341.x.
 |

