Biology:T-box leader
| T-box leader | |
|---|---|
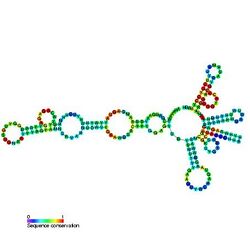 Predicted secondary structure and sequence conservation of T-box | |
| Identifiers | |
| Symbol | T-box |
| Rfam | RF00230 |
| Other data | |
| RNA type | Cis-reg; leader; |
| Domain(s) | Bacteria |
| GO | 0000049 |
| SO | 0000140 |
| PDB structures | PDBe |
Usually found in gram-positive bacteria, the T box leader sequence is an RNA element that controls gene expression through the regulation of translation by binding directly to a specific tRNA and sensing its aminoacylation state.[1] This interaction controls expression of downstream aminoacyl-tRNA synthetase genes, amino acid biosynthesis, and uptake-related genes in a negative feedback loop.[1][2] The uncharged tRNA acts as the effector for transcription antitermination of genes in the T-box leader family.[3][4][5] The anticodon of a specific tRNA base pairs to a specifier sequence within the T-box motif, and the NCCA acceptor tail of the tRNA base pairs to a conserved bulge in the T-box antiterminator hairpin.[6]
tRNA-mediated attenuation
Although the exact mechanism of T box leader is still unclear and currently being studied, it has recently been recognized as a member of an expanding group of RNAs that are phylogenetically conserved across many gram-positive bacteria.[2] They are structurally complex and able to directly sense physiological signals which results in the control of downstream gene expression.[2] This controlling of gene expression is accomplished by transcriptional attenuation—a general transcriptional regulation strategy that senses when an alteration in the rate of transcription is necessary and initiating alteration at a particular site (sometimes preceding one or more genes of an operon).[7] The operons that encode aminoacyl-tRNA synthetases, regulated by tRNA-mediated transcriptional attenuation, contain a leader region that specifies a transcript segment that can fold and eventually form a complex set of structures.[7] Two of the most crucial segments to attenuation function as both the terminator and the antiterminator in different regulatory situations.[7]
Leader structure
In terms of structure, the T box RNA is highly conserved—especially in the stem I distal region.[1] The stem I region forms an arched conformation, with the apex containing a complex loop-loop interaction between the conserved adenine-guanine bulge and distal loop.[1] This loop-loop structure is similar to that seen in the ribosome exit site, suggesting that it is highly conserved among tRNA recognition sites.[1] The apex of the stem I region recognizes two critical positions on the tRNA: the anticodon and D/T-loops.[8] Extensive intermolecular interactions occur at this site.[8] If the length or orientation of these two recognition points is altered or mismatched, the T box riboswitch and tRNA complex is disrupted, and proper functioning of transcriptional regulation cannot occur.[8][9]
Riboswitch function
The riboswitch functions by directly sensing a physiological signal.[10] Next, a specific uncharged tRNA binds to a riboswitch element in the transcript, and a structural change occurs in the transcript that promotes expression of the downstream coding sequence.[2][10] The specifier sequence is the first recognition sequence in the leader.[7] It is complementary to the anticodon of the tRNA that is a substrate of the tRNA synthetase under regulation.[7] The second tRNA binding sequence, the T box sequence, is complementary to the nucleotide preceding the acceptor end of the tRNA.[7] The T box is found in the side bulge of the antiterminator.[7]
Method of regulation
The most common model system used to study T-box leader is in the gram-positive bacterium Bacillus subtilis.[10] In terms of what is currently understood about the regulatory role of T box function, it appears that when the uncharged tRNA is abundant, it binds to the specifier and the T box sequence of an appropriate leader RNA, stabilizing the antiterminator and, in turn, preventing terminator formation.[7] Without terminator formation, transcription will proceed.[7] If, however, the tRNA is charged, its acceptor end will be blocked by an amino acid and thus, cannot pair with the T box.[7] The terminator will then form, thereby terminating transcription.[7]
External links
- Page for T-box leader at Rfam
- TBDB, a database of annotated T-box leader sequences
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "T box RNA decodes both the information content and geometry of tRNA to affect gene expression". Proceedings of the National Academy of Sciences of the United States of America 110 (18): 7240–7245. April 2013. doi:10.1073/pnas.1222214110. PMID 23589841. Bibcode: 2013PNAS..110.7240G.
- ↑ 2.0 2.1 2.2 2.3 "The T box mechanism: tRNA as a regulatory molecule". FEBS Letters 584 (2): 318–324. January 2010. doi:10.1016/j.febslet.2009.11.056. PMID 19932103.
- ↑ "Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base". Journal of Bacteriology 176 (15): 4518–4526. August 1994. doi:10.1128/jb.176.15.4518-4526.1994. PMID 8045882.
- ↑ "tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene". RNA 6 (8): 1131–1141. August 2000. doi:10.1017/s1355838200992100. PMID 10943892.
- ↑ "The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs". RNA 7 (8): 1165–1172. August 2001. doi:10.1017/s1355838201002370. PMID 11497434.
- ↑ "Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility". Journal of Molecular Biology 326 (1): 189–201. February 2003. doi:10.1016/s0022-2836(02)01339-6. PMID 12547201.
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 Lederberg, ed.-in-chief: Joshua (2000). Encyclopedia of microbiology (2. ed.). San Diego [u.a.]: Academic Press. ISBN 978-0-12-226800-7. https://archive.org/details/encyclopediaofmi02edunse.
- ↑ 8.0 8.1 8.2 "Structural determinants for geometry and information decoding of tRNA by T box leader RNA". Structure 21 (11): 2025–2032. November 2013. doi:10.1016/j.str.2013.09.001. PMID 24095061.
- ↑ "Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA". Journal of Molecular Biology 408 (1): 99–117. April 2011. doi:10.1016/j.jmb.2011.02.014. PMID 21333656.
- ↑ 10.0 10.1 10.2 Henkin, Tina. "Research Interests". http://microbiology.osu.edu/faculty/henkin-tina-m.
 |



