Biology:Dronpa
Dronpa is a reversibly switchable photoactivatable fluorescent protein that is 2.5 times as bright as EGFP.[1][2] Dronpa gets switched off by strong illumination with 488 nm (blue) light and this can be reversed by weak 405 nm UV light.[1] A single dronpa molecule can be switched on and off over 100 times.[3] It has an excitation peak at 503 nm and an emission peak at 518 nm.
History
A tetrameric,[4] reversibly switchable fluorescent protein was discovered in a cDNA screen of a stony coral (Pectiniidae). A monomeric variant of this protein was named "Dronpa" after "Dron" a ninja term for vanishing and pa for photoactivation.[2]
Structure and mechanism of photoswitching
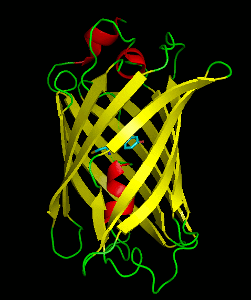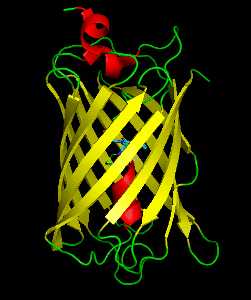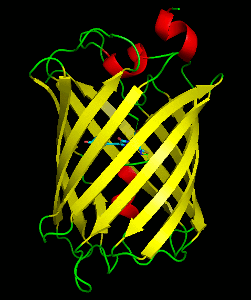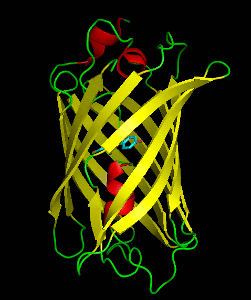
Dronpa is 257 amino acids long and is a 28.8 kDa monomer. Dronpa is 76% similar in sequence to GFP[2] and shares a similar structure with an 11 stranded β-barrel (a β-can) enclosing an α-helix.[5] The chromophore is formed autocatalytically from residues Cys62, Tyr63 and Gly64.[5][6] The on state of the dronpa molecule has the chromophore in a cis conformation while the off state chromophore exists in the trans conformation. Several other residues in the vicinity of the chromophore also move during the on-off transition resulting a very different electrostatic environment.[5]
Applications
Dronpa's fast dynamics and stability under repeated cycles of switching make it one of the more important switchable fluorescent proteins.[1] It is used in super resolution microscopy techniques like PALM/STORM. It can also be used to track fast dynamics of proteins in cells.
Oligomeric forms of Dronpa have been engineered as synthetic photosensory domains. When a dimeric or tetrameric form of Dronpa photoswitches, its oligomerization affinity changes. This was used to enable optical control over the activity of enzymes. Specifically, two Dronpa domains can be attached to locations on a protein so that their tetramerization or oligomerization blocks or cages protein function in the dark, but monomerization after illumination activates or uncages protein function. This method has been used to control a variety of proteins including serine/threonine kinases.[4][7]
References
- ↑ 1.0 1.1 1.2 Day, R. N.; Davidson, M. W. (2009). "The fluorescent protein palette: Tools for cellular imaging". Chemical Society Reviews 38 (10): 2887–2921. doi:10.1039/b901966a. PMID 19771335.
- ↑ 2.0 2.1 2.2 Ando, R.; Mizuno, H.; Miyawaki, A. (2004). "Regulated Fast Nucleocytoplasmic Shuttling Observed by Reversible Protein Highlighting". Science 306 (5700): 1370–1373. doi:10.1126/science.1102506. PMID 15550670. Bibcode: 2004Sci...306.1370A.
- ↑ Habuchi, S. (2005). "Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa". Proceedings of the National Academy of Sciences 102 (27): 9511–9516. doi:10.1073/pnas.0500489102. PMID 15972810. Bibcode: 2005PNAS..102.9511H.
- ↑ 4.0 4.1 Zhou, X. X.; Chung, H. K.; Lam, A. J.; Lin, M. Z. (2012). "Optical Control of Protein Activity by Fluorescent Protein Domains". Science 338 (6108): 810–814. doi:10.1126/science.1226854. PMID 23139335. Bibcode: 2012Sci...338..810Z.
- ↑ 5.0 5.1 5.2 Andresen, M.; Stiel, A. C.; Trowitzsch, S.; Weber, G.; Eggeling, C.; Wahl, M. C.; Hell, S. W.; Jakobs, S. (2007). "Structural basis for reversible photoswitching in Dronpa". Proceedings of the National Academy of Sciences 104 (32): 13005–9. doi:10.1073/pnas.0700629104. PMID 17646653. Bibcode: 2007PNAS..10413005A.
- ↑ Trowitzsch, S.; Stiel, A. C.; Weber, G.; Andresen, M.; Eggeling, C.; Hell, S. W.; Jakobs, S.; Wahl, M. C. (2007). "1.8 Å bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants". Biochemical Journal 402 (1): 35–42. doi:10.1042/BJ20061401. PMID 17117927.
- ↑ Zhou, X. X.; Fan, L. Z.; Li, P.; Shen, K.; Lin, M.Z. (2017). "Optical control of cell signaling by single-chain photoswitchable kinases". Science 355 (6327): 836–842. doi:10.1126/science.aah3605. PMID 28232577. Bibcode: 2017Sci...355..836Z.
 |