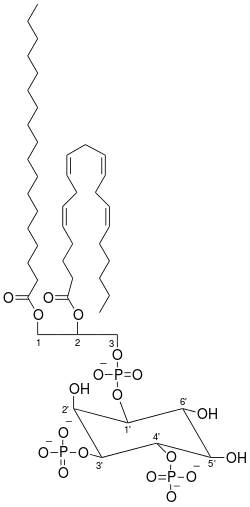
Chemistry:Phosphatidylinositol 3,4-bisphosphate
Phosphatidylinositol (3,4)-bisphosphate (PtdIns(3,4)P2) is a minor phospholipid component of cell membranes, yet an important second messenger. The generation of PtdIns(3,4)P2 at the plasma membrane activates a number of important cell signaling pathways.[1]
Of all the phospholipids found within the membrane, inositol phospholipids make up less than 10%.[2] Phosphoinositide’s (PI’s), also known as phosphatidylinositol phosphates, are synthesized in the cell's endoplasmic reticulum by the protein phosphatidylinositol synthase (PIS).[3][4][5] PI’s are highly compartmentalized; their main components include a glycerol backbone, two fatty acid chains enriched with stearic acid and arachidonic acid, and an inositol ring whose phosphate groups' regulation differs between organelles depending on the specific PI and PIP kinases and PIP phosphatases present in the organelle.[6][7][8] These kinases and phosphatases conduct phosphorylation and dephosphorylation at the inositol sugar head groups 3’, 4’, and 5’ positions, producing differing phosphoinositides, including PtdIns(3,4)P2.[9][1] PI kinases catalyze phosphate groups binding while PI phosphatases remove phosphate groups at the three positions on the PI inositol ring, giving seven different combinations of PI’s.[10][11]
PtdIns(3,4)P2 is dephophosphorylated by the phosphatase INPP4B on the 4' position of the inositol ring and by the TPTE (transmembrane phosphatases with tensin homology) family of phosphatases on the 3 position of the inositol ring.
The PH domain in a number of proteins binds to PtdIns(3,4)P2 including the PH domain in PKB. The generation of PtdIns(3,4)P2 at the plasma membrane upon the activation of class I PI 3-kinases and SHIP phosphatases causes these proteins to translocate to the plasma membrane, thereby affecting their activity.
Class I and II phosphoinositide 3-kinases (PI3Ks) synthesize PtdIns(3,4)P2 by phosphorylating the phosphoinositide PI4P’s 3' -OH position.[12][13] Phosphatases SHIP1 and SH2-containing inositol 5’-polyphosphatases (SHIP2) produce PtdIns(3,4)P2 through desphosphorylation of PtdIns(3,4,5)P3’s 5’ inositol ring position.[14][15] In addition to these positive regulators at the plasma membrane (PM), 3-phosphatase tensin homolog (PTEN) acts as a negative regulator of PtdIns(3,4)P2 production by depleting PtdIns(3,4,5)P3 levels at the PM through dephosphorylation of PtdIns(3,4,5)P3’s 3’ inositol ring position, giving rise to PtdIns(4,5)P2.[16][17] Inositol polyphosphate 4-phosphatase isozymes, INPP4A and INPP4B, also act as negative PtdIns(3,4)P2 regulators, though through a more direct interaction- by hydrolyzing PtdIns(3,4)P2’s 4-phosphate, producing PI3P.[18][19][20] PtdIns(3,4)P2 has been indicated to be critical for AKT (Protein kinase B, PKB) activation within the PI3K pathway through the PI’s regulation by the SHIP1 and 2 phosphatases. Akt is recruited and subsequently activated through its PH domains interaction with PtdIns(3,4)P2 and PtdIns(3,4,5)P3 both of which have shown to have high affinity with the Akt PH domain.[21] Once bound to the PM through its interaction with PtdIns(3,4)P2 and PtdIns(3,4,5)P3, Akt is activated through release of its auto-inhibitory interaction between the PH and kinase domains.[22] Following this release, T308 in the proteins activation loop and S437 in the proteins hydrophobic domain are phosphorylated by Phosphoinositide-dependent kinase-1 (PDK1)[23] and mechanistic target of Rapamycin Complex 2 (mTORC2),[24] respectively. Test tube experiments have shown that the essential recruitment of PDK1 for Akt activation at the PM can be driven through interactions with both PtdIns(3,4)P2 and PtdIns(3,4,5)P3.[25]
It was originally presumed that 5-phosphatases dephosphorylation of PI(3,4,5)P3 would be anti-tumoral, similar to tumor suppressor PTEN. Yet the 5-phosphatase SHIP proteins synthesis of PI(3,4)P2 has been linked to tumor cell survival due to the lipid’s binding and subsequent activation of Akt.[26] Akt activation causes downstream metabolism alterations, apoptosis suppression and a rise in cell proliferation.[27] This pathway and its effects have shown up in 50% of cancers.[28] In conjunction, investigators have shown a rise in PI(3,4)P2 levels and mutation of 4-phosphatase INPP4B has shown mammary epithelial transformation.[29]
Recently, PtdIns(3,4)P2 has been shown to play an important role in vesicle maturation during clathrin-mediated endocytosis (CME).[30][31] PtdIns(4)P synthesizing phosphatases SHIP2 and synaptojanin are recruited to clathrin structures at the beginning of the CME process.[32][33] This production of PtdIns(4)P subsequently leads to PtdIns(3,4)P2 synthesis through PI3K-C2α11, and the newly synthesized PtdIns(3,4)P2 then recruits SNX9 and SNX18 PX-BAR domain proteins which narrow the nascent vesicles neck to eventually be cut and released by dynamin, forming vesicles.[34][35]
PI(3,4)P2 plays another possible role at the PM, promoting cytoskeletal rearrangements through actin regulatory proteins like Lamellipodin.[36][37] Lamellipodin is recruited to the PM where it is believed to interact with PI(3,4)P2 through its PH domain. Once at the PM, it can regulate lamellipodia actin networks and cell migration by interacting with actin-binding proteins like Ena/VASP.[38][39][40]
References
- ↑ Dimitrios Karathanassis; Robert V. Stahelin; Jerónimo Bravo; Olga Perisic; Christine M Pacold; Wonhwa Cho; Roger L Williams (2002). "Binding of the PX domain of p47phox to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction". EMBO Journal 21 (19): 5057–5068. doi:10.1093/emboj/cdf519. PMID 12356722.
- ↑ Alberts, B., Johnson, A., Lewis, J., Morgan, D., Raff, M., Roberts, K., & Walters, P. (2015). Molecular biology of the cell (Sixth ed.). New York, NY: Garland Science.
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Agranoff BW, Bradley R M, Brady RO. The enzymatic synthesis of inositol phosphatide. J Biol Chem. (1958) 233:1077–83.
- ↑ Epand RM. Recognition of polyunsaturated acyl chains by enzymes acting on membrane lipids. Biochim Biophys Acta. (2012) 1818:957–62. 10.1016/j.bbamem.2011.07.018
- ↑ Agranoff BW, Bradley R M, Brady RO. The enzymatic synthesis of inositol phosphatide. J Biol Chem. (1958) 233:1077–83.
- ↑ Epand RM. Recognition of polyunsaturated acyl chains by enzymes acting on membrane lipids. Biochim Biophys Acta. (2012) 1818:957–62. 10.1016/j.bbamem.2011.07.018
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Alberts, B., Johnson, A., Lewis, J., Morgan, D., Raff, M., Roberts, K., & Walters, P. (2015). Molecular biology of the cell (Sixth ed.). New York, NY: Garland Science.
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. (2013) 93:1019–137. 10.1152/physrev.00028.2012
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. (2013) 93:1019–137. 10.1152/physrev.00028.2012
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Fernandes S, Iyer S, Kerr WG. Role of SHIP1 in cancer and mucosal inflammation. Ann NY Acad Sci. (2013) 1280:6–10. 10.1111/nyas.12038
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Kerr WG. Inhibitor and activator: dual functions for SHIP in immunity and cancer. Ann NY Acad Sci. (2011) 1217:1–17. 10.1111/j.1749-6632.2010.05869.x
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Norris FA, Atkins RC, Majerus PW. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. evidence for conserved alternative splicing in the 4-phosphatase family. J Biol Chem. (1997) 272:23859–64. 10.1074/jbc.272.38.23859
- ↑ Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. . Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. (2009) 16:115–25. 10.1016/j.ccr.2009.06.006
- ↑ Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA. High Affinity Binding of Inositol Phosphates and Phosphoinositides to the Pleckstrin Homology Domain of RAC/Protein Kinase B and Their Influence on Kinase Activity. The Journal of biological chemistry. 1997;272(13):8474–8481.
- ↑ Ebner M, Lučić I, Leonard TA, Yudushkin I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell. 2017;65(3):416-431.e6.
- ↑ Alessi DR, James SR, Downes CP, Holmes AB, Gaffney P, Reese CB, et al. Characterization of a 3- phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Current Biology. 1997;7(4).
- ↑ Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR Complex. Science. 2005;307(5712):1098–101.
- ↑ Alessi DR, James SR, Downes CP, Holmes AB, Gaffney P, Reese CB, et al. Characterization of a 3- phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Current Biology. 1997;7(4).
- ↑ Ebner M, Lučić I, Leonard TA, Yudushkin I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell. 2017;65(3):416-431.e6.
- ↑ Ebner M, Lučić I, Leonard TA, Yudushkin I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell. 2017;65(3):416-431.e6.
- ↑ Ebner M, Lučić I, Leonard TA, Yudushkin I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol Cell. 2017;65(3):416-431.e6.
- ↑ Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. . Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. (2009) 16:115–25. 10.1016/j.ccr.2009.06.006
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, Ullrich A, Lampe A, et al. . Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. (2013) 499:233–7. 10.1038/nature12360
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Nakatsu F, Perera RM, Lucast L, Zoncu R, Domin J, Gertler FB, et al. . The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J Cell Biol. (2010) 190:307–15. 10.1083/jcb.201005018
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, Ullrich A, Lampe A, et al. . Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. (2013) 499:233–7. 10.1038/nature12360
- ↑ Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., & Martini, M. (2020). PI(3,4)P2 Signaling in Cancer and Metabolism. Frontiers in oncology, 10, 360. https://doi.org/10.3389/fonc.2020.00360
- ↑ Hawkins PT, Stephens LR. Emerging evidence of signalling roles for PI(3,4)P2 in class I and II PI3K-regulated pathways. Biochem Soc Trans. (2016) 44:307–14. 10.1042/BST20150248
- ↑ Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, et al. . Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. (2004) 7:571–83. 10.1016/j.devcel.2004.07.024 [PubMed] [CrossRef] [Google Scholar]
- ↑ Yoshinaga S, Ohkubo T, Sasaki S, Nuriya M, Ogawa Y, Yasui M, et al. . A phosphatidylinositol lipids system, lamellipodin, and Ena/VASP regulate dynamic morphology of multipolar migrating cells in the developing cerebral cortex. J Neurosci. (2012) 32:11643–56. 10.1523/JNEUROSCI.0738-12.2012 [PMC free article] [PubMed] [CrossRef] [Google Scholar]
- ↑ (22)Kato T, Kawai K, Egami Y, Kakehi Y, Araki N. Rac1-dependent lamellipodial motility in prostate cancer PC-3 cells revealed by optogenetic control of Rac1 activity. PLoS ONE. (2014) 9:e97749. 10.1371/journal.pone.0097749
 |