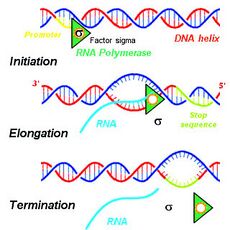
Biology:Termination signal
A termination signal is a sequence that signals the end of transcription or translation.[1] Termination signals are found at the end of the part of the chromosome being transcribed during transcription of mRNA. Termination signals bring a stop to transcription, ensuring that only gene-encoding parts of the chromosome are transcribed.[1] Transcription begins at the promoter when RNA polymerase, an enzyme that facilitates transcription of DNA into mRNA, binds to a promoter, unwinds the helical structure of the DNA, and uses the single-stranded DNA as a template to synthesize RNA.[1] Once RNA polymerase reaches the termination signal, transcription is terminated.[1] In bacteria, there are two main types of termination signals: intrinsic and factor-dependent terminators.[1] In the context of translation, a termination signal is the stop codon on the mRNA that elicits the release of the growing peptide from the ribosome.[2]
Termination signals play an important role in regulating gene expression since they mark the end of a gene transcript and determine which DNA sequences are expressed in the cell.[1] Expression levels of certain genes can be increased by inhibiting signal terminators, known as antitermination, which allows for transcription to continue beyond the termination signal site.[1] This can be desirable under specific cell conditions.[1]
Additionally, sometimes, termination signals are overlooked in transcription and translation, resulting in unwanted transcription or translation past the termination signal.[3] To address this issue, termination signals can be optimized to increase termination efficiency.[3]
Bacterial Termination Signals
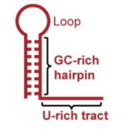
The two types of termination signals in bacteria are intrinsic and factor-dependent terminators.[4] Intrinsic termination occurs when a specific sequence on the growing RNA strand elicits detachment of RNA polymerase from the RNA-DNA complex.[4] In E. coli, one intrinsic termination signal consists of an RNA hairpin that has high amounts of guanine and cytosine, as well as a region high in uracil nucleobases.[4]
Factor-dependent terminators require proteins for proper termination.[4] One example is rho-dependent termination, a common termination mechanism found in bacteria that involves the binding of Rho protein to remove RNA polymerase from the DNA-RNA complex.[4]
Antitermination
Antitermination involves the inhibition of signal terminators.[4] RNA polymerase is prevented from detaching from the RNA in response to a termination signal, increasing downstream gene expression.[4]
Antitermination can occur in a variety of ways.[4] Some antiterminators disrupt termination signals by inhibiting RNA hairpin generation, while other antiterminators are proteins that bind to RNA polymerase and cause RNA polymerase to continue transcription past termination signals.[4] Depending on the environment of the cell, antitermination may be crucial to cell survival.[4] These antitermination mechanisms are crucial when the cell is under stress, allowing for increased expression of downstream genes that are needed under dire circumstances.[4]
Termination Signal Efficiency
Transcription
Termination efficiency of T7 RNA polymerase is around 74%, which creates issues when T7 RNA polymerase is used to produce recombinant proteins.[3] In this process, the target gene is inserted into a plasmid and is regulated by the T7 promoter; T7 RNA polymerase is used to transcribe the target gene.[3] Due to termination inefficiency, read-through can result in increased regulation of downstream genes that may be crucial to host cell function.[3] Selectable marker genes that are downstream of the target gene insertion site and genes that encode regulatory proteins may have altered expression as a result.[3] Hence, proper termination of transcription is needed for plasmid stability in host cells for the proper production of recombinant proteins.[3] Research has been conducted to identify termination signals that yield higher termination efficiency by engineering termination signals from a variety of termination signal components.[3] Some engineered termination signals have yielded a termination efficiency as high as 99%, which is a significant improvement from the native termination efficiency associated with T7 RNA polymerase of 74%.[3]
Translation
In translation, termination efficiency is dependent on the context of the termination signal (stop codon).[2] Traditionally, the termination signal for translation is a 3 nucleobase sequence called a stop codon.[2] Research has shown that the nucleobases surrounding the stop codon can impact termination efficiency.[2] Specifically, the 4th base (nucleobase directly following the stop codon) has a significant impact on the termination efficiency.[2] In particular, when the nucleobase at the 4th position is a purine (adenine or guanine), termination efficiency is improved.[2] Pyrimidines (cytosine or uracil) in the 4th position result in lower termination efficiency.[2] It has been found that highly expressed genes have higher termination efficiency due to the presence of a purine in the 4th position.[2]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Pelley, John W. (2012-01-01), Pelley, John W., ed., "16 – RNA Transcription and Control of Gene Expression" (in en), Elsevier's Integrated Review Biochemistry (Second Edition) (Philadelphia: W.B. Saunders): pp. 137–147, ISBN 978-0-323-07446-9, https://www.sciencedirect.com/science/article/pii/B9780323074469000167, retrieved 2021-10-15
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 McCaughan, K. K.; Brown, C. M.; Dalphin, M. E.; Berry, M. J.; Tate, W. P. (1995-06-06). "Translational termination efficiency in mammals is influenced by the base following the stop codon" (in en). Proceedings of the National Academy of Sciences of the United States of America 92 (12): 5431–5435. doi:10.1073/pnas.92.12.5431. ISSN 0027-8424. PMID 7777525. Bibcode: 1995PNAS...92.5431M.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Mairhofer, Juergen; Wittwer, Alexander; Cserjan-Puschmann, Monika; Striedner, Gerald (2015-03-20). "Preventing T7 RNA Polymerase Read-through Transcription—A Synthetic Termination Signal Capable of Improving Bioprocess Stability". ACS Synthetic Biology 4 (3): 265–273. doi:10.1021/sb5000115. PMID 24847676. https://doi.org/10.1021/sb5000115.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 Santangelo, Thomas; Artsimovitch, Irina (9 May 2011). "Termination and antitermination: RNA polymerase runs a stop sign". Nature Reviews Microbiology 9 (5): 319–329. doi:10.1038/nrmicro2560. PMID 21478900.
- Merrill, Dr. Gary F. 'Transcription', lecture notes distributed in Biochemistry 451 General Biochemistry, Oregon State University, Weigend on 6 June 2006.
 |