Biology:Sulfoglycolysis
Sulfoglycolysis is a catabolic process in primary metabolism in which sulfoquinovose (6-deoxy-6-sulfonato-glucose) is metabolized to produce energy and carbon-building blocks.[1][2] Sulfoglycolysis pathways occur in a wide variety of organisms, and enable key steps in the degradation of sulfoquinovosyl diacylglycerol (SQDG), a sulfolipid found in plants and cyanobacteria into sulfite and sulfate. Sulfoglycolysis converts sulfoquinovose (C6H12O8S−) into various smaller metabolizable carbon fragments such as pyruvate and dihydroxyacetone phosphate that enter central metabolism. The free energy is used to form the high-energy molecules ATP (adenosine triphosphate) and NADH (reduced nicotinamide adenine dinucleotide). Unlike glycolysis, which allows metabolism of all carbons in glucose, sulfoglycolysis pathways convert only a fraction of the carbon content of sulfoquinovose into smaller metabolizable fragments; the remainder is excreted as C3-sulfonates 2,3-dihydroxypropanesulfonate (DHPS) or sulfolactate (SL); or C2-sulfonates isethionate or sulfoacetate.
Several sulfoglycolytic pathways are known:
- The sulfoglycolytic Embden-Meyerhof-Parnas (sulfo-EMP) pathway, first identified in Escherichia coli, involves the degradation of sulfoquinovose to 2,3-dihydroxypropanesulfonate (DHPS),[3] and shares similarity with the Embden-Meyerhof-Parnas glycolysis pathway. This pathway leads to the production of the C3 intermediate dihydroxyacetone phosphate.
- The sulfoglycolytic Entner-Doudoroff (sulfo-ED) pathway, first identified in Pseudomonas putida SQ1, involves the degradation of sulfoquinovose to sulfolactate,[4] and shares similarity to the Entner-Doudoroff pathway of glycolysis. This pathway leads to the production of the C3 intermediate pyruvate.
- The sulfofructose transaldolase pathway, first identified in Bacillus aryabhattai [5] and Bacillus megaterium,[6] involves isomerization of SQ to sulfofructose, and then a transaldolase cleaves SF to 3-sulfolactaldehyde (SLA), while the non-sulfonated C3-(glycerone)-moiety is transferred to an acceptor molecule, glyceraldehyde phosphate (GAP), yielding fructose-6-phosphate (F6P). The SLA released can either be oxidized (to sulfolactate) or reduced (to dihydroxypropanesulfonate) and then excreted.
- The sulfoglycolytic transketolase (sulfo-TL) pathway was first identified in Clostridium sp. MSTE9.[7] It involves isomerization of SQ to sulfofructose, and then a transketolase cleaves SF to 4-sulfoerythrose (SE), while the C2-moiety is transferred to an acceptor molecule, glyceraldehyde phosphate (GAP), yielding xylulose-5-phosphate (Xu5P). 4-Sulfoerythrose is isomerized to 4-sulfoerythrulose (SEu), whereupon a second round of transketolase catalyzed reaction cleaves SE to sulfoacetaldehyde, while the non-sulfonated C2-moiety is transferred to an acceptor molecule, glyceraldehyde phosphate (GAP), yielding a second molecule of xylulose-5-phosphate (Xu5P). Finally, the sulfoacetaldehyde is reduced to isethionate and excreted.
Additionally, there are sulfoquinovose 'sulfolytic' pathways that allow degradation of sulfoquinovose through cleavage of the C-S bond. These include:
- The sulfoglycolytic sulfoquinovose monooxygenase (sulfo-SMO) pathway, first identified in Agrobacterium tumerfaciens [8] and Novosphingobium aromaticivorans,[9] involves the degradation of sulfoquinovose to glucose and sulfite. Glucose formed in this pathway enters glycolysis.
- The sulfoglycolytic sulfoquinovose dioxygenase (sulfo-SMO) pathway.
In all pathways, energy is formed by breakdown of the carbon-rich fragments in later stages through the 'pay-off' phase of glycolysis through substrate-level phosphorylation to produce ATP and NADH.
Growth of bacteria on sulfoquinovose and its glycosides
A range of bacteria can grow on sulfoquinovose or its glycosides as sole carbon source. E. coli can grow on sulfoquinovose,[3] methyl α-sulfoquinovoside and α-sulfoquinovosyl glycerol.[10] Growth on sulfoquinovosyl glycerol is both faster and leads to higher cell density than for growth on sulfoquinovose.[10] Pseudomonas aeruginosa strain SQ1,[11] Klebsiella sp. strain ABR11,[12] Klebsiella oxytoca TauN1,[11] Agrobacterium sp. strain ABR2,[12] and Bacillus aryabhattai[5] can grow on sulfoquinovose as sole carbon source. A strain of Flavobacterium was identified that could grow on methyl α-sulfoquinovoside.[13]
Production of sulfoquinovose and its mutarotation
Sulfoquinovose is rarely found in its free form in nature; rather it occurs predominantly as a glycoside, SQDG. SQDG can be deacylated to form lyso-SQDG and sulfoquinovosylglycerol (SQGro).[14][15][16] Sulfoquinovose is obtained from SQ glycosides by the action of sulfoquinovosidases, which are glycoside hydrolases that can hydrolyse the glycosidic linkage in SQDG, or its deacylated form, sulfoquinovosyl glycerol (SQGro).[17]
There are two main classes of sulfoquinovosidases. The first are classical glycoside hydrolases (which belong to CAZy family GH31), and is exemplified by the sulfoquinovosidase YihQ from Escherichia coli. Family GH31 sulfoquinovosidases cleave SQ glycosides with retention of configuration, initially forming α-sulfoquinovose. YihQ sulfoquinovosidase exhibits a preference for the naturally occurring 2’R-SQGro.[10] The second class of sulfoquinovosidases are NAD+-dependent enzymes (which belong to CAZy family GH188) that use an oxidoreductive mechanism to cleave both α- and β-glycosides of sulfoquinovose.[18]
Sulfoglycolysis encoding operons often contain gene sequences encoding aldose-1-epimerases that act as sulfoquinovose mutarotases, catalyzing the interconversion of the α and β anomers of sulfoquinovose.[19]
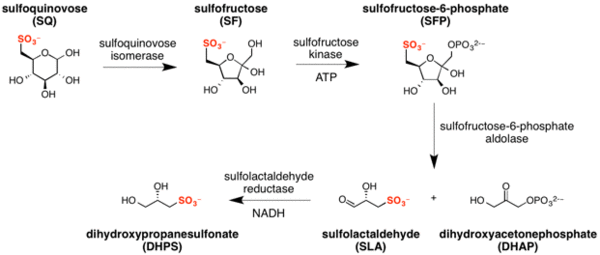
Sulfo-EMP pathway
The major steps in the sulfo-EMP pathway[3] are:
- isomerization of sulfoquinovose to sulfofructose (catalyzed by sulfoquinovose isomerase), with transient formation of sulforhamnose;[20]
- phosphorylation of sulfofructose to sulfofructose-1-phosphate (catalyzed by sulfofructose kinase and using ATP as a co-factor);
- retro-aldol cleavage of sulfofructose-1-phosphate to afford dihydroxyacetone phosphate and (S)-sulfolactaldehyde (catalyzed by sulfofructose-1-phosphate aldolase);
- reduction of sulfolactaldehyde to (S)-2,3-dihydroxypropane-1-sulfonate (catalyzed by sulfolactaldehyde reductase and using NADH as a co-factor).[21]
Expression of proteins within the sulfo-EMP operon of E. coli is regulated by a transcription factor termed CsqR (formerly YihW).[22] CsqR binds to DNA sites within the operon encoding the sulfo-EMP pathway, functioning as a repressor. SQ, SQGro and the transiently formed intermediate sulforhamnose (but not lactose, glucose or galactose) function as derepressors of CsqR.[20]
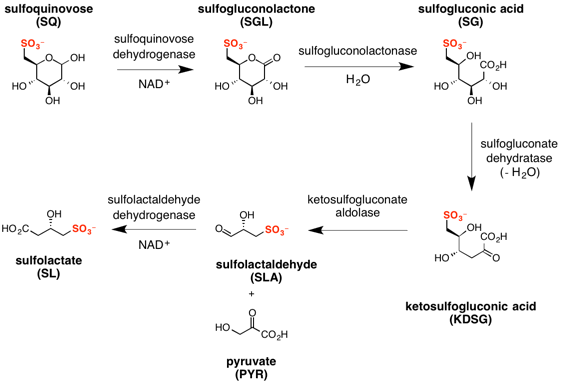
Sulfo-ED pathway
The major steps in the sulfo-ED pathway[4] are:
- oxidation of sulfoquinovose to sulfogluconolactone (catalyzed by sulfoquinovose dehydrogenase with NAD+ co-factor);
- hydrolysis of sulfogluconolactone to sulfogluconate acid (catalyzed by sulfogluconolactonase with water);
- dehydration of sulfogluconic acid to 2-keto-3,6-dideoxy-6-sulfogluconate (catalyzed by sulfogluconate dehydratase);
- retro-aldol cleavage of 2-keto-3,6-dideoxy-6-sulfogluconate to give pyruvate and (S)-sulfolactaldehyde (catalyzed by sulfoketogluconate dehydrogenase with NAD+ co-factor);
- oxidation of sulfolactaldehyde to (S)-sulfolactate (catalyzed by sulfolactaldehyde dehydrogenase with NAD+ co-factor).
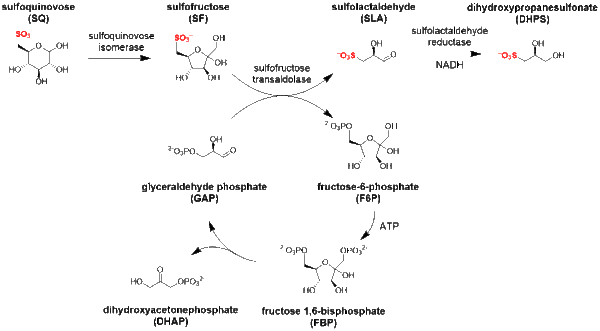
Sulfo-SFT pathway
The major steps in the sulfo-SFT pathway[5] are:
- isomerization of sulfoquinovose to sulfofructose (catalyzed by sulfoquinovose isomerase);
- transaldol reaction of sulfofructose to release sulfolactaldehyde (catalyzed by sulfofructose transaldolase), and transfer of the C3-(glycerone)-moiety to glyceraldehyde phosphate, yielding fructose-6-phosphate;
- sulfolactaldehyde may be reduced to (S)-2,3-dihydroxypropane-1-sulfonate (catalyzed by sulfolactaldehyde reductase and using NADH as a co-factor), or oxidized to sulfolactate (catalyzed by sulfolactaldehyde dehydrogenase using NAD+ as a co-factor).
The transaldolase can also catalyze transfer of the C3-(glycerone)-moiety to erythrose-4-phosphate, giving sedoheptulose-7-phosphate.
Sulfo-TK pathway
The major steps in the Sulfo-TK pathway[23] are:
- isomerization of sulfoquinovose to sulfofructose (catalyzed by sulfoquinovose isomerase);
- transketol reaction of sulfofructose to release erythrose (catalyzed by sulfofructose transketolase, a thiamine diphosphate dependent enzyme), and transfer of the C2-moiety to glyceraldehyde phosphate, yielding xylulose-5-phosphate (Xu5P).
- 4-Sulfoerythrose is isomerized to 4-sulfoerythrulose (SEu), whereupon a second round of transketolase catalyzed reaction cleaves SE to sulfoacetaldehyde, while the C2-moiety is again transferred to an acceptor molecule, glyceraldehyde phosphate (GAP), yielding a second molecule of xylulose-5-phosphate (Xu5P).
- Finally, the sulfoacetaldehyde is reduced to isethionate and excreted.
The sulfoacetaldehyde may be oxidized to sulfoacetate.
Degradation of DHPS and SL
The C3 sulfonates DHPS and SL are metabolized for their carbon content, as well as to mineralize their sulfur content.[2] Metabolism of DHPS typically involves conversion to SL. Metabolism of SL can occur in several ways including:
- elimination of sulfite to afford pyruvate;
- oxidation to sulfopyruvate, transamination to cysteate, and elimination of sulfite to afford pyruvate and ammonia;
- oxidation to sulfopyruvate, decarboxylation to sulfoacetaldehyde, and phosphorylation to afford acetylphosphate and sulfite.
See also
References
- ↑ Snow, Alexander J. D.; Burchill, Laura; Sharma, Mahima; Davies, Gideon J.; Williams, Spencer J. (2021). "Sulfoglycolysis: catabolic pathways for metabolism of sulfoquinovose". Chemical Society Reviews 50 (24): 13628–13645. doi:10.1039/D1CS00846C. PMID 34816844. https://eprints.whiterose.ac.uk/180655/1/CS_SYN_09_2021_000846.R2_Proof_hi.pdf.
- ↑ 2.0 2.1 "Sulfoquinovose in the biosphere: occurrence, metabolism and functions". The Biochemical Journal 474 (5): 827–849. February 2017. doi:10.1042/BCJ20160508. PMID 28219973.
- ↑ 3.0 3.1 3.2 "Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle". Nature 507 (7490): 114–7. March 2014. doi:10.1038/nature12947. PMID 24463506. Bibcode: 2014Natur.507..114D. http://nbn-resolving.de/urn:nbn:de:bsz:352-260772.
- ↑ 4.0 4.1 "Entner-Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1". Proceedings of the National Academy of Sciences of the United States of America 112 (31): E4298–305. August 2015. doi:10.1073/pnas.1507049112. PMID 26195800. Bibcode: 2015PNAS..112E4298F.
- ↑ 5.0 5.1 5.2 Frommeyer, B; Fiedler, AW; Oehler, SR; Hanson, BT; Loy, A; Franchini, P; Spiteller, D; Schleheck, D (28 August 2020). "Environmental and Intestinal Phylum Firmicutes Bacteria Metabolize the Plant Sugar Sulfoquinovose via a 6-Deoxy-6-sulfofructose Transaldolase Pathway.". iScience 23 (9): 101510. doi:10.1016/j.isci.2020.101510. PMID 32919372. Bibcode: 2020iSci...23j1510F.
- ↑ Liu, Y; Wei, Y; Zhou, Y; Ang, EL; Zhao, H; Zhang, Y (17 December 2020). "A transaldolase-dependent sulfoglycolysis pathway in Bacillus megaterium DSM 1804.". Biochemical and Biophysical Research Communications 533 (4): 1109–1114. doi:10.1016/j.bbrc.2020.09.124. PMID 33036753.
- ↑ Liu, Jiayi; Wei, Yifeng; Ma, Kailiang; An, Junwei; Liu, Xumei; Liu, Yinbo; Ang, Ee Lui; Zhao, Huimin et al. (17 December 2021). "Mechanistically Diverse Pathways for Sulfoquinovose Degradation in Bacteria". ACS Catalysis 11 (24): 14740–14750. doi:10.1021/acscatal.1c04321.
- ↑ Sharma, M; Lingford, JP; Petricevic, M; Snow, AJD; Zhang, Y; Järvå, MA; Mui, JW; Scott, NE et al. (25 January 2022). "Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.". Proceedings of the National Academy of Sciences of the United States of America 119 (4): e2116022119. doi:10.1073/pnas.2116022119. PMID 35074914. Bibcode: 2022PNAS..11916022S.
- ↑ Liu, Jiayi; Wei, Yifeng; Ma, Kailiang; An, Junwei; Liu, Xumei; Liu, Yinbo; Ang, Ee Lui; Zhao, Huimin et al. (17 December 2021). "Mechanistically Diverse Pathways for Sulfoquinovose Degradation in Bacteria". ACS Catalysis 11 (24): 14740–14750. doi:10.1021/acscatal.1c04321.
- ↑ 10.0 10.1 10.2 Abayakoon, Palika; Jin, Yi; Lingford, James P.; Petricevic, Marija; John, Alan; Ryan, Eileen; Wai-Ying Mui, Janice; Pires, Douglas E.V. et al. (2018-09-05). "Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases" (in en). ACS Central Science 4 (9): 1266–1273. doi:10.1021/acscentsci.8b00453. ISSN 2374-7943. PMID 30276262.
- ↑ 11.0 11.1 "Sulfoquinovose degraded by pure cultures of bacteria with release of C3-organosulfonates: complete degradation in two-member communities". FEMS Microbiology Letters 328 (1): 39–45. March 2012. doi:10.1111/j.1574-6968.2011.02477.x. PMID 22150877.
- ↑ 12.0 12.1 "Glycolytic breakdown of sulfoquinovose in bacteria: a missing link in the sulfur cycle". Applied and Environmental Microbiology 69 (11): 6434–41. November 2003. doi:10.1128/AEM.69.11.6434-6441.2003. PMID 14602597. Bibcode: 2003ApEnM..69.6434R.
- ↑ "Sulfocarbohydrate metabolism. I. bacterial production and utilization of sulfoacetate". Biochimica et Biophysica Acta 93: 169–71. October 1964. doi:10.1016/0304-4165(64)90272-7. PMID 14249144.
- ↑ "Metabolism of sulfoquinovosyl diglyceride in Chlorella pyrenoidosa by sulfoquinovosyl monoglyceride: fatty acyl CoA acyltransferase and sulfoquinovosyl glyceride: fatty acyl ester hydrolase pathways". Journal of Lipid Research 15 (1): 1–10. January 1974. doi:10.1016/S0022-2275(20)36825-5. PMID 4359538.
- ↑ "Metabolism of the plant sulfolipid--sulfoquinovosyldiacylglycerol: degradation in animal tissues". Archives of Biochemistry and Biophysics 259 (2): 510–9. December 1987. doi:10.1016/0003-9861(87)90517-0. PMID 3426241.
- ↑ "Hydrolysis of galactolipids by human pancreatic lipolytic enzymes and duodenal contents". Journal of Lipid Research 36 (6): 1392–400. June 1995. doi:10.1016/S0022-2275(20)41146-0. PMID 7666015.
- ↑ "YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids". Nature Chemical Biology 12 (4): 215–7. April 2016. doi:10.1038/nchembio.2023. PMID 26878550. http://eprints.whiterose.ac.uk/98363/1/compiled_YihQ_final_submission_for_GJD.pdf.
- ↑ Kaur, Arashdeep; Pickles, Isabelle B.; Sharma, Mahima; Madeido Soler, Niccolay; Scott, Nichollas E.; Pidot, Sacha J.; Goddard-Borger, Ethan D.; Davies, Gideon J. et al. (27 December 2023). "Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism". Journal of the American Chemical Society 145 (51): 28216–28223. doi:10.1021/jacs.3c11126.
- ↑ "Discovery and characterization of a sulfoquinovose mutarotase using kinetic analysis at equilibrium by exchange spectroscopy". The Biochemical Journal 475 (7): 1371–1383. April 2018. doi:10.1042/BCJ20170947. PMID 29535276.
- ↑ 20.0 20.1 Sharma, Mahima; Abayakoon, Palika; Epa, Ruwan; Jin, Yi; Lingford, James P.; Shimada, Tomohiro; Nakano, Masahiro; Mui, Janice W.-Y. et al. (23 February 2021). "Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis". ACS Central Science 7 (3): 476–487. doi:10.1021/acscentsci.0c01285. PMID 33791429.
- ↑ Sharma, Mahima; Abayakoon, Palika; Lingford, James P.; Epa, Ruwan; John, Alan; Jin, Yi; Goddard-Borger, Ethan D.; Davies, Gideon J. et al. (21 February 2020). "Dynamic Structural Changes Accompany the Production of Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase". ACS Catalysis 10 (4): 2826–2836. doi:10.1021/acscatal.9b04427. https://eprints.whiterose.ac.uk/156563/1/acscatal.9b04427.pdf.
- ↑ Shimada, Tomohiro; Yamamoto, Kaneyoshi; Nakano, Masahiro; Watanabe, Hiroki; Schleheck, David; Ishihama, Akira (29 October 2018). "Regulatory role of CsqR (YihW) in transcription of the genes for catabolism of the anionic sugar sulfoquinovose (SQ) in Escherichia coli K-12". Microbiology 165 (1): 78–89. doi:10.1099/mic.0.000740. PMID 30372406.
- ↑ Liu, Jiayi; Wei, Yifeng; Ma, Kailiang; An, Junwei; Liu, Xumei; Liu, Yinbo; Ang, Ee Lui; Zhao, Huimin et al. (17 December 2021). "Mechanistically Diverse Pathways for Sulfoquinovose Degradation in Bacteria". ACS Catalysis 11 (24): 14740–14750. doi:10.1021/acscatal.1c04321.
 |