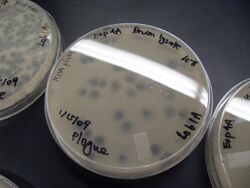
Biology:Viral plaque
A viral plaque is a visible structure formed after introducing a viral sample to a cell culture grown on some nutrient medium. The virus will replicate and spread, generating regions of cell destruction known as plaques. For example, Vero cell or other tissue cultures may be used to investigate an influenza virus or coronavirus, while various bacterial cultures would be used for bacteriophages.[1][2]
Counting the number of plaques can be used as a method of virus quantification. These plaques can sometimes be detected visually using colony counters, in much the same way as bacterial colonies are counted; however, they are not always visible to the naked eye, and sometimes can only be seen through a microscope, or using techniques such as staining (e.g. neutral red for eukaryotes[3] or giemsa for bacteria[4]) or immunofluorescence. Special computer systems have been designed with the ability to scan samples in batches.
The appearance of the plaque depends on the host strain, virus and the conditions. Highly virulent or lytic strains create plaques that look clear (due to total cell destruction), while strains that only kill a fraction of their hosts (due to partial resistance/lysogeny), or only reduce the rate of cell growth, give turbid plaques. Some partially lysogenic phages give bull's-eye plaques with spots or rings of growth in the middle of clear regions of complete lysis.[5]
See also
References
- ↑ Abedon, Stephen T. (2018), Harper, David R.; Abedon, Stephen T.; Burrowes, Benjamin H. et al., eds., "Detection of Bacteriophages: Phage Plaques" (in en), Bacteriophages: Biology, Technology, Therapy (Cham: Springer International Publishing): pp. 1–32, doi:10.1007/978-3-319-40598-8_16-1, ISBN 978-3-319-40598-8, https://doi.org/10.1007/978-3-319-40598-8_16-1, retrieved 2023-12-02
- ↑ Youil, R; Su, Q; Toner, T. J; Szymkowiak, C; Kwan, W-S; Rubin, B; Petrukhin, L; Kiseleva, I et al. (2004-09-01). "Comparative study of influenza virus replication in Vero and MDCK cell lines". Journal of Virological Methods 120 (1): 23–31. doi:10.1016/j.jviromet.2004.03.011. ISSN 0166-0934. PMID 15234806. https://www.sciencedirect.com/science/article/pii/S0166093404001016.
- ↑ Finter, N. B (1969-10-01). "Dye Uptake Methods for Assessing Viral Cytopathogenicity and Their Application to Interferon Assays". Journal of General Virology 5 (3): 419–427. doi:10.1099/0022-1317-5-3-419. ISSN 0022-1317. http://vir.sgmjournals.org/content/5/3/419. Retrieved 2012-03-26.
- ↑ Marvin, D. A.; Hohn, B. (1969). "Filamentous bacterial viruses". Bacteriological Reviews 33 (2): 172–209. doi:10.1128/br.33.2.172-209.1969. PMID 4979697.
- ↑ Ramesh, Nachimuthu; Archana, Loganathan; Madurantakam Royam, Madhav; Manohar, Prasanth; Eniyan, Kandasamy (2019-06-17). "Effect of various bacteriological media on the plaque morphology of Staphylococcus and Vibrio phages". Access Microbiology 1 (4): e000036. doi:10.1099/acmi.0.000036. ISSN 2516-8290. PMID 32974524.
External links
- An image of Bacteriophage plaques in agar
 |