Biology:Chromogenic in situ hybridization
Chromogenic in situ hybridization (CISH) is a cytogenetic technique that combines the chromogenic signal detection method of immunohistochemistry (IHC) techniques with in situ hybridization.[1][2] It was developed around the year 2000 as an alternative to fluorescence in situ hybridization (FISH) for detection of HER-2/neu oncogene amplification.[1] CISH is similar to FISH in that they are both in situ hybridization techniques used to detect the presence or absence of specific regions of DNA.[1] However, CISH is much more practical in diagnostic laboratories because it uses bright-field microscopes rather than the more expensive and complicated fluorescence microscopes used in FISH.[1][3]
Procedure
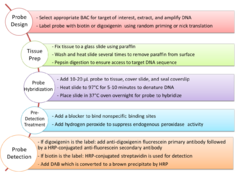
Probe design
Probe design for CISH is very similar to that for FISH with differences only in labelling and detection. FISH probes are generally labelled with a variety of different fluorescent tags and can only be detected under a fluorescence microscope,[4] whereas CISH probes are labelled with biotin or digoxigenin [5] and can be detected using a bright-field microscope after other treatment steps have been applied.[1]
CISH probes are approximately 20 nucleotides in length and are designed for DNA targets. They are complementary to the targeted sequence and bind to it after a denaturation and hybridization step. Only a few CISH probes are available commercially, so for most applications they have to be extracted, amplified, sequenced, labelled and mapped from bacterial artificial chromosomes (BACs).[6] BACs were developed during the Human Genome Project as it was necessary to isolate and amplify short fragments of human DNA for sequencing purposes.[7] Nowadays, BACs can be selected and positioned on the human genome using public databases such as the UCSC Genome Browser.[6] This ensures optimal complementarity and sequence specificity. DNA is extracted from the BAC clones and amplified using a polymerase-based technique, such as degenerate oligonucleotide primed (DOP)-PCR.[8] Next, the clones are sequenced and their position on the genome is verified.[9] Probe labelling can be carried out by using either random priming or nick translation to incorporate biotin or digoxigenin.[10]
Preparation of tissue, hybridization of probes, and detection
For CISH to work optimally, chromosomes must be in either interphase or metaphase. Tissue samples are securely attached to a surface, which is usually a glass slide, with paraffin.[11] The tissue samples must then be washed and heated several times to remove any paraffin before the hybridization step. After this, the sample has to undergo pepsin digestion to ensure the target is accessible.[11] As a final step, 10–20 μL of probe is added, the sample is covered with a coverslip which is sealed with rubber cement, and the slide is heated to 97 °C for 5–10 minutes to denature the DNA.[11] The slide is then placed in a 37 °C oven overnight so that the probe can hybridize.[11][12] On the next day, the sample is washed and a blocker for nonspecific protein binding sites is applied.[11] If horseradish peroxidase (HRP) is going to be used, the sample must be incubated in hydrogen peroxide to suppress endogenous peroxidase activity.[11] If digoxigenin was used as a probe label, an anti-digoxigenin fluorescein primary antibody followed by a HRP-conjugated anti-fluorescein secondary antibody are then applied.[1] If biotin was used as a probe label, non-specific binding sites must first be blocked using bovine serum albumin (BSA).[11] Then, HRP-conjugated streptavidin is used for detection.[6][11] HRP then converts diaminobenzidine (DAB) into an insoluble brown product, which can be detected in a bright-field microscope under 40- to 60-fold magnification.[11][13] Staining lightly with hematoxylin can be used as a counterstain to make the product more visible.[5]
Comparison to other techniques
Compared to FISH
FISH is considered to be the gold standard for the detection of chromosomal abnormalities because it is very sensitive and has high resolution.[3][14] Other techniques that are developed to detect chromosomal abnormalities are usually compared to the sensitivity and specificity of FISH to see how they measure up.[3][14] For example, compared to FISH, CISH has been shown to have a sensitivity of 97.5% and a specificity of 94% for detection of HER-2/neu gene amplification.[3] The concordance rate between FISH and CISH was 94.8%, showing CISH to be a comparable technique to FISH.[3] Most other sources agree and report an almost equal performance on gene amplification assays for FISH and CISH.[15][16] However, sometimes CISH shows lower sensitivity for low level amplifications.[1]
CISH has some advantages over FISH in the reagents and equipment it uses. As noted above, CISH is much cheaper and is easier to use because it uses bright-field microscopes instead of fluorescence microscopes.[1][3] In addition, the CISH reagents are more stable than the FISH reagents so it is possible to store the samples and examine the same sample multiple times.[3][14] FISH reagents fade over time due to photobleaching so a sample can only be examined once.[3][14] Apart from the expensive fluorescence microscope, FISH also requires a high-resolution digital camera to capture micrographs of the sample before the fluorescence fades.[14] Another advantage of using bright-field microscopy is that the tissue or cell sample as a whole can be visualized through CISH whereas cell morphology is difficult to assess using fluorescence microscopy in FISH.[3][14]
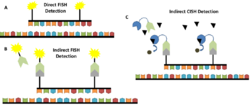
CISH also differs from FISH in the probes that are used as well as in the overall method. There are many different types of FISH probes available, such as repeat probes, probes that detect specific genes or telomeres, and probes that detect chromosomal abnormalities.[14] In contrast, there is a limited variety of commercially available CISH probes, including probes that bind the centromere of chromosomes 3, 7, 8, 9, 10, 11, 17, 18, X, and Y as well as gene-specific probes for cancer-related genes, such as HER-2, EGFR, MYC, and TOP2A.[14] Despite the limited variety of available CISH probes, they are generally more cost-effective than FISH probes.[14] With regard to the overall method, FISH can be performed using direct labelling—fluorochromes are attached to the probes—or indirect labelling—the probes are labelled with biotin or digoxigenin which are then detected using fluorescently-labelled streptavidin or antibodies, respectively.[14] CISH is performed using indirect labelling in which antibodies or streptavidin are conjugated to enzymes such as HRP or alkaline phosphatase (AP).[14]
Compared to IHC
CISH and IHC are similar in that both are used for the same purpose (mainly to detect HER-2/neu amplification) and they both use enzyme reactions (HRP/AP) to measure amplification.[13] CISH and IHC are different in that IHC measures protein expression whereas CISH measures DNA amplification.[13] This difference is particularly useful for HER-2/neu because it has been found that gene amplification is of higher prognostic value than protein expression.[17]
A disadvantage of IHC is that it is not possible to identify false-negative and false-positive results.[17] In CISH, if there is no signal for the reference probe, the assay has failed.
For low and high protein overexpression/gene amplification, CISH and IHC show a concordance of over 86% and over 89%, respectively.[18] It has been shown that monoclonal antibodies are better than polyclonal antibodies for detection in both IHC and CISH as they bind more specifically, which leads to a higher concordance rate.[18]
For medium protein overexpression/gene amplification concordance varies, but is higher when monoclonal antibodies are used than when polyclonal antibodies are used.[18] The variable concordance is due to the fact that gene amplification does not strictly correlate with protein expression and that tumor heterogeneity can make it difficult to detect protein overexpression in a tissue.[18]
Medical applications
CISH is frequently applied to assess gene amplification, such as HER-2/neu status in breast cancer samples.[2][19] HER-2/neu amplification is associated with higher mortality, higher recurrence rate, and poor prognosis in breast cancer.[20] The monoclonal antibody trastuzumab is a receptor blocker that has been proven to be clinically very effective in HER-2/neu-overexpressing tumors.[21] Therefore, it is crucial to determine receptor status before starting cancer treatment.[21]
CISH is also used for detection of chromosomal rearrangements and fusions, such as the fusion of ALK tyrosine kinase domain with the promoter and 5’ region of EML4 in lung cancer. ALK-positive tumors are a clinically relevant subgroup as they can be very effectively treated with the ALK inhibitor crizotinib.[22][23]
Apart from cancers, CISH has also been shown to be useful in detecting human papillomavirus infections.[24]
Variations
Silver-enhanced in situ hybridization (SISH)
SISH uses a similar method as CISH, but a silver precipitate is the end product rather than a brown product.[25] In this method, a probe tagged with dinitrophenol (DNP) binds to the target sequence.[25] A primary anti-DNP antibody is then added followed by a secondary antibody conjugated to HRP.[25] Silver acetate, hydroquinone, and hydrogen peroxide are subsequently added to the secondary antibody and HRP catalyzes the polymerization of silver in the presence of hydrogen peroxide.[25] Silver metal is consequently deposited into the nuclei of the cell.[25] Amplification of HER-2/neu is seen as black dots.[25]
DuoCISH
DuoCISH is a variation of CISH that addresses the need for two different probes on the same slide.[26] It is a well-established technique for HER-2/neu amplification detection even though it is sometimes reported to be less effective than FISH.[27] In this technique, one probe binds the reference which, in the case of HER-2/neu amplification detection, is CEN17 (the centromere of chromosome 17) while the other probe binds the target sequence which is HER-2/neu.[26][27] DuoCISH combines both FISH and CISH in that it converts signals from FISH probes to chromogenic substrates.[26][27] It works on the principle that blue cyanine dye is a substrate for HRP and red dye is a substrate for AP. For example, green fluorescein isothiocyanate (FITC) signals are converted to blue chromogenic precipitates through an anti-FITC antibody conjugated to HRP, and Texas Red signals are converted to red chromogenic precipitates through an anti-Texas Red antibody conjugated to AP.[26][27] An advantage of DuoCISH is that it is possible to distinguish between chromosomal aneuploidy and gene amplification as the reference gene will also be amplified in aneuploidy but not in gene amplification detection.[26][27]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Tanner, M; Gancberg, D; Di Leo, A; Larsimont, D; Rouas, G; Piccart, M. J.; Isola, J (2000). "Chromogenic in situ hybridization: A practical alternative for fluorescence in situ hybridization to detect HER-2/neu oncogene amplification in archival breast cancer samples". The American Journal of Pathology 157 (5): 1467–72. doi:10.1016/S0002-9440(10)64785-2. PMID 11073807.
- ↑ 2.0 2.1 Madrid, M. A.; Lo, R. W. (2004). "Chromogenic in situ hybridization (FISH): A novel alternative in screening archival breast cancer tissue samples for HER-2/neu status". Breast Cancer Research 6 (5): R593–600. doi:10.1186/bcr915. PMID 15318940.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Sáez, A; Andreu, F. J.; Seguí, M. A.; Baré, M. L.; Fernández, S; Dinarés, C; Rey, M (2006). "HER-2 gene amplification by chromogenic in situ hybridisation (CISH) compared with fluorescence in situ hybridisation (FISH) in breast cancer-A study of two hundred cases". The Breast 15 (4): 519–27. doi:10.1016/j.breast.2005.09.008. PMID 16290155.
- ↑ Garimberti, E; Tosi, S (2010). "Fluorescence in situ Hybridization (FISH), Basic Principles and Methodology". Fluorescence in situ Hybridization (FISH). Methods in Molecular Biology. 659. pp. 3–20. doi:10.1007/978-1-60761-789-1_1. ISBN 978-1-60761-788-4.
- ↑ 5.0 5.1 Lambros, M. B.; Simpson, P. T.; Jones, C; Natrajan, R; Westbury, C; Steele, D; Savage, K; MacKay, A et al. (2006). "Unlocking pathology archives for molecular genetic studies: A reliable method to generate probes for chromogenic and fluorescent in situ hybridization". Laboratory Investigation 86 (4): 398–408. doi:10.1038/labinvest.3700390. PMID 16446704.
- ↑ 6.0 6.1 6.2 Summersgill, B. M.; Shipley, J. M. (2010). "Fluorescence in Situ Hybridization Analysis of Formalin Fixed Paraffin Embedded Tissues, Including Tissue Microarrays". Fluorescence in situ Hybridization (FISH). Methods in Molecular Biology. 659. pp. 51–70. doi:10.1007/978-1-60761-789-1_4. ISBN 978-1-60761-788-4.
- ↑ Shizuya, H; Kouros-Mehr, H (2001). "The development and applications of the bacterial artificial chromosome cloning system". The Keio Journal of Medicine 50 (1): 26–30. doi:10.2302/kjm.50.26. PMID 11296661.
- ↑ Darouich, S; Popovici, C; Missirian, C; Moncla, A (2012). "Use of DOP-PCR for amplification and labeling of BAC DNA for FISH". Biotechnic & Histochemistry 87 (2): 117–21. doi:10.3109/10520295.2011.559175. PMID 21314248.
- ↑ De Braekeleer, E; Douet-Guilbert, N; Basinko, A; Morel, F; Le Bris, M. J.; Férec, C; De Braekeleer, M (2011). "Using bacterial artificial chromosomes in leukemia research: The experience at the university cytogenetics laboratory in Brest, France". Journal of Biomedicine and Biotechnology 2011: 1–7. doi:10.1155/2011/329471. PMID 21274439.
- ↑ Cold Spring Harbor Laboratory Press. (2012) "Labeling of DNA Probes by Nick Translation". Molecular Cloning: A Laboratory Manual.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 11.8 Park, K; Kim, J; Lim, S; Han, S; Lee, J. Y. (2003). "Comparing fluorescence in situ hybridization and chromogenic in situ hybridization methods to determine the HER2/neu status in primary breast carcinoma using tissue microarray". Modern Pathology 16 (9): 937–43. doi:10.1097/01.MP.0000086487.78558.7D. PMID 13679458.
- ↑ Schildhaus, H. U.; Deml, K. F.; Schmitz, K; Meiboom, M; Binot, E; Hauke, S; Merkelbach-Bruse, S; Büttner, R (2013). "Chromogenic in situ hybridization is a reliable assay for detection of ALK rearrangements in adenocarcinomas of the lung". Modern Pathology 26 (11): 1468–77. doi:10.1038/modpathol.2013.95. PMID 23743932.
- ↑ 13.0 13.1 13.2 Gupta, D; Middleton, L. P.; Whitaker, M. J.; Abrams, J (2003). "Comparison of fluorescence and chromogenic in situ hybridization for detection of HER-2/neu oncogene in breast cancer". American Journal of Clinical Pathology 119 (3): 381–7. doi:10.1309/P40P2EAD42PUKDMG. PMID 12645340.
- ↑ 14.00 14.01 14.02 14.03 14.04 14.05 14.06 14.07 14.08 14.09 14.10 Lambros, M. B.; Natrajan, R; Reis-Filho, J. S. (2007). "Chromogenic and fluorescent in situ hybridization in breast cancer". Human Pathology 38 (8): 1105–22. doi:10.1016/j.humpath.2007.04.011. PMID 17640550.
- ↑ Zhao, J; Wu, R; Au, A; Marquez, A; Yu, Y; Shi, Z (2002). "Determination of HER2 gene amplification by chromogenic in situ hybridization (CISH) in archival breast carcinoma". Modern Pathology 15 (6): 657–65. doi:10.1038/modpathol.3880582. PMID 12065780.
- ↑ Shia, J; Klimstra, D. S.; Li, A. R.; Qin, J; Saltz, L; Teruya-Feldstein, J; Akram, M; Chung, K. Y. et al. (2005). "Epidermal growth factor receptor expression and gene amplification in colorectal carcinoma: An immunohistochemical and chromogenic in situ hybridization study". Modern Pathology 18 (10): 1350–6. doi:10.1038/modpathol.3800417. PMID 15832190.
- ↑ 17.0 17.1 Todorović-Raković, N; Jovanović, D; Nesković-Konstantinović, Z; Nikolić-Vukosavljević, D (2005). "Comparison between immunohistochemistry and chromogenic in situ hybridization in assessing HER-2 status in breast cancer". Pathology International 55 (6): 318–23. doi:10.1111/j.1440-1827.2005.01831.x. PMID 15943788.
- ↑ 18.0 18.1 18.2 18.3 Rosa, F. E.; Santos, R. M.; Rogatto, S. R.; Domingues, M. A. (2013). "Chromogenic in situ hybridization compared with other approaches to evaluate HER2/neu status in breast carcinomas". Brazilian Journal of Medical and Biological Research 46 (3): 207–16. doi:10.1590/1414-431x20132483. PMID 23558859.
- ↑ Kumamoto, H; Sasano, H; Taniguchi, T; Suzuki, T; Moriya, T; Ichinohasama, R (2001). "Chromogenic in situ hybridization analysis of HER-2/neu status in breast carcinoma: Application in screening of patients for trastuzumab (Herceptin) therapy". Pathology International 51 (8): 579–84. doi:10.1046/j.1440-1827.2001.01255.x. PMID 11564211.
- ↑ Mitri, Z; Constantine, T; O'Regan, R (2012). "The HER2 Receptor in Breast Cancer: Pathophysiology, Clinical Use, and New Advances in Therapy". Chemotherapy Research and Practice 2012: 1–7. doi:10.1155/2012/743193. PMID 23320171.
- ↑ 21.0 21.1 Swain, S. M.; Kim, S. B.; Cortés, J; Ro, J; Semiglazov, V; Campone, M; Ciruelos, E; Ferrero, J. M. et al. (2013). "Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): Overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study". The Lancet Oncology 14 (6): 461–71. doi:10.1016/S1470-2045(13)70130-X. PMID 23602601.
- ↑ Kwak, E. L.; Bang, Y. J.; Camidge, D. R.; Shaw, A. T.; Solomon, B; Maki, R. G.; Ou, S. H.; Dezube, B. J. et al. (2010). "Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer". New England Journal of Medicine 363 (18): 1693–703. doi:10.1056/NEJMoa1006448. PMID 20979469.
- ↑ Wagner, F; Streubel, A; Roth, A; Stephan-Falkenau, S; Mairinger, T (2014). "Chromogenic in situ hybridisation (CISH) is a powerful method to detect ALK-positive non-small cell lung carcinomas". Journal of Clinical Pathology 67 (5): 403–7. doi:10.1136/jclinpath-2013-201974. PMID 24293609.
- ↑ Zappacosta, R; Colasante, A; Viola, P; d'Antuono, T; Lattanzio, G; Capanna, S; Gatta, D. M.; Rosini, S (2013). "Chromogenic in situ hybridization and p16/Ki67 dual staining on formalin-fixed paraffin-embedded cervical specimens: Correlation with HPV-DNA test, E6/E7 mRNA test, and potential clinical applications". BioMed Research International 2013: 1–11. doi:10.1155/2013/453606. PMID 24369532.
- ↑ 25.0 25.1 25.2 25.3 25.4 25.5 Shousha, S; Peston, D; Amo-Takyi, B; Morgan, M; Jasani, B (2009). "Evaluation of automated silver-enhanced in situ hybridization (SISH) for detection of HER2 gene amplification in breast carcinoma excision and core biopsy specimens". Histopathology 54 (2): 248–53. doi:10.1111/j.1365-2559.2008.03185.x. PMID 19207950.
- ↑ 26.0 26.1 26.2 26.3 26.4 Henriksen, U., Müller, S., and Schønau, A. (2009) Chapter 14: "Dual color CISH and FISH to CISH conversion", pp. 97–101 in G.L. Kumar and L. Rudbeck (Eds.), Immunohistochemical (IHC) Staining Methods Fifth Edition. Carpinteria, CA: Dako North America
- ↑ 27.0 27.1 27.2 27.3 27.4 Mollerup, J; Henriksen, U; Müller, S; Schønau, A (2012). "Dual color chromogenic in situ hybridization for determination of HER2 status in breast cancer: A large comparative study to current state of the art fluorescence in situ hybridization". BMC Clinical Pathology 12: 3. doi:10.1186/1472-6890-12-3. PMID 22333181.
 |