Biology:Ivy-DE RNA motif
| ivy-DE | |
|---|---|
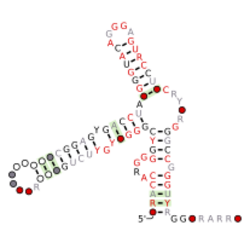 Consensus secondary structure and sequence conservation of ivy-DE RNA | |
| Identifiers | |
| Symbol | ivy-DE |
| Rfam | RF02999 |
| Other data | |
| RNA type | Cis-reg |
| SO | 0005836 |
| PDB structures | PDBe |
The ivy-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] ivy-DE motifs are found in the genus Pseudomonas.
ivy-DE motif RNAs likely function as cis-regulatory elements, in view of their positions that are consistently downstream of protein-coding genes. However, the RNA is most likely located in the 3′ UTR of the regulated (upstream) genes, even though cis-regulatory RNAs in bacteria generally reside in the 5′ UTR. No genetic elements are consistently located downstream of ivy-DE motif RNAs, so the RNAs could be regulators within the 3′ UTR. However, it is possible that they are co-transcribed with the upstream genes and function rather as small RNAs.
ivy-DE motif RNAs are downstream of ivy (inhibitor of vertebrate lysozyme) genes. ivy proteins are used by bacteria and have shown to be potent inhibitors of vertebrate lysozymes (see [1]). However, other biological functions have been proposed.[2] Additionally, at least in Pseudomonas aeruginosa PAO1, the RNA is associated with a gene that encodes a homolog of the ivy protein that does not actually inhibit lysozyme.[2] The function of this gene is unknown, but its expression increases in bacterial strains that overexpress the PhrS small RNA[3] and during hypoxic growth.[4]
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ 2.0 2.1 "The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase". J. Biol. Chem. 285 (20): 14843–14847. May 2010. doi:10.1074/jbc.C110.120931. PMID 20351104.
- ↑ "The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal". Mol. Microbiol. 80 (4): 868–885. May 2011. doi:10.1111/j.1365-2958.2011.07620.x. PMID 21375594.
- ↑ "Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration". Mol. Microbiol. 65 (1): 153–165. July 2007. doi:10.1111/j.1365-2958.2007.05772.x. PMID 17581126.
 |

