Biology:DNase footprinting assay
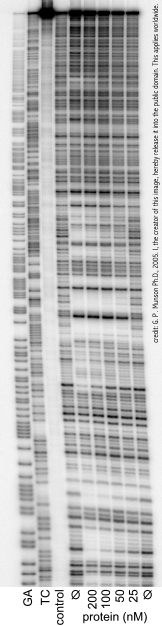
A DNase footprinting assay[1] is a DNA footprinting technique from molecular biology/biochemistry that detects DNA-protein interaction using the fact that a protein bound to DNA will often protect that DNA from enzymatic cleavage. This makes it possible to locate a protein binding site on a particular DNA molecule. The method uses an enzyme, deoxyribonuclease (DNase, for short), to cut the radioactively end-labeled DNA, followed by gel electrophoresis to detect the resulting cleavage pattern.
For example, the DNA fragment of interest may be PCR amplified using a 32P 5' labeled primer, with the result being many DNA molecules with a radioactive label on one end of one strand of each double stranded molecule. Cleavage by DNase will produce fragments. The fragments which are smaller with respect to the 32P-labelled end will appear further on the gel than the longer fragments. The gel is then used to expose a special photographic film.
The cleavage pattern of the DNA in the absence of a DNA binding protein, typically referred to as free DNA, is compared to the cleavage pattern of DNA in the presence of a DNA binding protein. If the protein binds DNA, the binding site is protected from enzymatic cleavage. This protection will result in a clear area on the gel which is referred to as the "footprint".
By varying the concentration of the DNA-binding protein, the binding affinity of the protein can be estimated according to the minimum concentration of protein at which a footprint is observed.
This technique was developed by David J. Galas and Albert Schmitz at Geneva in 1977[2]
See also
- DNA footprinting
- DNase II
- Toeprinting assay
References
- ↑ "[9] Quantitative DNase footprint titration: A method for studying protein-DNA interactions". Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods in Enzymology. 130. 1986. pp. 132–81. doi:10.1016/0076-6879(86)30011-9. ISBN 9780121820305.
- ↑ "DNAse footprinting: a simple method for the detection of protein-DNA binding specificity". Nucleic Acids Research 5 (9): 3157–70. Sep 1978. doi:10.1093/nar/5.9.3157. PMID 212715.
 |