Biology:HOLDH RNA motif
| HOLDH | |
|---|---|
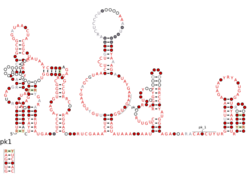 Consensus secondary structure and sequence conservation of HOLDH RNA | |
| Identifiers | |
| Symbol | HOLDH |
| Rfam | RF02997 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The Human, Oral, Large, Distant to HINT RNA motif (also HOLDH RNA motif) is a conserved RNA structure that was discovered by bioinformatics.[1] HOLDH motif RNAs are found exclusively in metagenomic sequences, especially those derived from human supragingival dental plaque and on the tongue.
HOLDH RNAs likely function in trans as small RNAs. In comparison to other bacterial small RNAs, they are located relatively far from the neighboring protein-coding genes (at least hundreds of base pairs). This extra non-coding sequence could form additional structure of HOLDH RNAs, but it is uncertain whether or not this is the case. The genes nearby to HOLDH RNAs are extremely similar, suggesting that the sequences are very closely related, which can hamper efforts to predict a conserved secondary structure using covariation (Biology). A gene encoding the HINT protein domain is consistently located nearby to HOLDH RNAs. However, given the high similarity between available examples of the HOLDH RNA motif, it is unclear how significant the HINT association is.
HOLDH RNAs contain a predicted kink turn.[2] This particular example of a kink turn was studied to better understand how kink turn structures relate to their sequences.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ "Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules". Nucleic Acids Res 49 (10): 5916–5924. June 2021. doi:10.1093/nar/gkab333. PMID 33978763.
 |

