Biology:Anti-hemB RNA motif
| anti-hemB | |
|---|---|
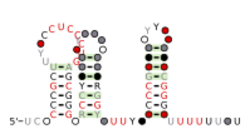 Consensus secondary structure and sequence conservation of Burkholderia RNA 7 (anti-hemB) | |
| Identifiers | |
| Symbol | anti-hemB |
| Rfam | RF02837 |
| Other data | |
| RNA type | Gene; sRNA |
| GO | 0005515,0097311 |
| SO | 0000370 |
| PDB structures | PDBe |
The anti-hemB RNA motif is a conserved RNA structure that was found in all known bacteria in the genus Burkholderia, and in a variety of other betaproteobacteria.[1] The anti-hemB RNA motif consists primarily of two stem-loops, followed by a predicted rho-independent transcription termination stem-loop. As anti-hemB RNAs are generally not located in a 5' UTR, the RNAs are presumed to be non-coding RNAs. The terminator stem-loop implies that anti-hemB RNAs are transcribed as independent molecules.
The name "anti-hemB" is to distinguish the motif from its reverse complement, which was called the "hemB" motif, a motif that is often in the apparent 5' UTRs of hemB genes. It was considered ambiguous as to which of these orientations is correct, as both directions exhibit covariation (see secondary structure prediction), and both fold into plausible RNA structures. However, the anti-hemB was judged the more likely orientation, because it had fewer A-C base pairs, which do not contribute to RNA stability. In reverse complement, an A-C pair becomes the stable G-U wobble pair.
References
- ↑ "Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline". Nucleic Acids Res. 35 (14): 4809–4819. 2007. doi:10.1093/nar/gkm487. PMID 17621584.
 |

