Biology:Chou's distorted key theory for peptide drugs
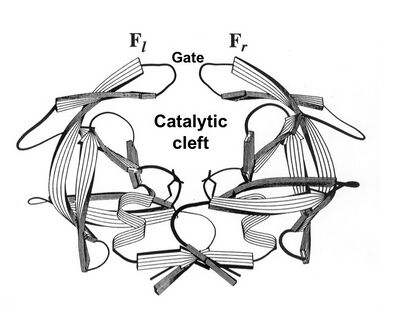
As a culprit of AIDS,[1][2] HIV protease has been a target for developing drugs against AIDS.[3][4] Functioning as a dimer of two identical subunits, HIV protease has a crab-like shape (Fig.1). Its catalytic cleft is gated by a pair of flaps (or pincers if viewed as a crab). When the enzyme is in an inhibitor-free state, the pincer-gate is open, allowing substrates to enter the catalytic cleft (Fig.1); when in an inhibitor-binding state, the pincer-gate is closed, blocking the entrance.
File:K2 HIV 8-site-binding.tiff
As a member of the aspartyl proteases, HIV protease is highly substrate-selective and cleavage-specific. Its susceptible sites in a protein extend to an octapeptide region (Fig.2) .[5]
Chou’s distorted key theory
Knowledge of the protein cleavage sites by HIV protease can provide very useful information for finding effective inhibitors against the culprit enzyme, as elaborated by Kuo-Chen Chou.[6] According to Fisher's lock-and-key model proposed by Hermann Emil Fischer in 1884 and Koshland's induced fit theory by Daniel E. Koshland, Jr. in 1958, given a peptide, the prerequisite condition for it to be cleaved by HIV-protease is a good fit and binding between the substrate and the enzyme's active site. However, such a peptide, after a modification on its scissile bond with some simple chemical procedure, will completely lose its cleavability but it can still tightly bind to the enzyme's active site. According to Kuo-Chen Chou, the molecule thus modified can be likened to a ‘‘distorted key”,[6] which can be inserted into a lock but can neither open the lock nor automatically get out from it. That is why a molecule modified from a cleavable peptide can spontaneously become a competitive inhibitor against the enzyme.
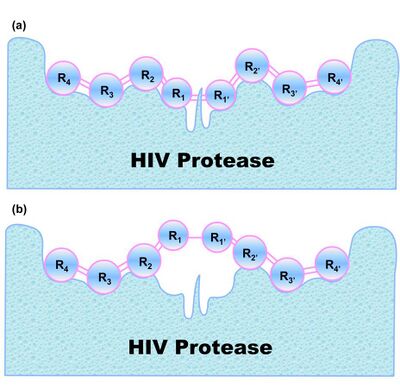
An illustration about Chou's distorted key theory is given in Fig.3, where panel (a) shows an effective binding of a cleavable peptide to the active site of HIV protease, while panel (b) the peptide has become a non-cleavable one after its scissile bond is modified although it can still bind to the active site. Such a modified peptide, or ‘‘distorted key”, will automatically become an inhibitor candidate against HIV protease.
Even for non-peptide inhibitors, the information derived from the cleavable peptides can also provide useful insights about the key binding groups and fitting conformation, among many other detailed requirements in microenvironment.
Many efforts have been made to predict the protein cleavage sites by HIV-protease (see, e.g., [7] [8] [9] [10] Also, a web-server named HIVcleave was established [11] for predicting HIV protease cleavage sites in proteins.
Application
Chou's distorted key theory was also utilized to develop inhibitors against severe acute respiratory syndrome or SARS (see, e.g., [12] [13] [14]).
References
- ↑ Barre-Sinoussi, F.; Chermann, J. C.; Rey, F.; Nugeyre, M. T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C. et al. (1997). "Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS)". Science 220 (4599): 868–871. doi:10.1126/science.6189183. PMID 6189183.
- ↑ Gallo, R. C. et al. (1984). "Frequent detection". Science 224 (4648): 500–503. doi:10.1126/science.6200936. PMID 6200936.
- ↑ Meek, T. D. et al. (1990). "Inhibition of HIV-1 protease in infected T-lymphocytes by synthetic peptide analogues.". Nature 343 (1): 90–92. doi:10.1038/343090a0. PMID 1688646.
- ↑ McQuade, T. J.; Tomasselli, A. G.; Liu, L.; Karacostas, V.; Moss, B.; Sawyer, T. K.; Heinrikson, R. L.; Tarpley, W. G. (1990). "A synthetic HIV-1 protease inhibitor with antiviral activity arrests HIV-like particle maturation.". Science 247 (1): 454–456. doi:10.1126/science.2405486. PMID 2405486.
- ↑ Miller, M.; Schneider, J.; Sathyanarayana, B. K.; Toth, M. V.; Marshall, G. R.; Clawson, L.; Selk, L.; Kent, S. B. et al. (1989). "Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution.". Science 246 (4934): 1149–1152. doi:10.1126/science.2686029. PMID 2686029.
- ↑ Jump up to: 6.0 6.1 Chou K.C. (1996). "Review: Prediction of human immunodeficiency virus protease cleavage sites in proteins". Analytical Biochemistry 233: 1–14. doi:10.1006/abio.1996.0001. PMID 8789141.
- ↑ Chou, K.C. (August 1993). "A vectorized sequence-coupling model for predicting HIV protease cleavage sites in proteins". Journal of Biological Chemistry 268 (23): 16938–16948. PMID 8349584.
- ↑ Chou, J.J. (August 1993). "Predicting cleavability of peptide sequences by HIV protease via correlation-angle approach". Journal of Protein Chemistry 12 (3): 291–302. doi:10.1007/bf01028191. PMID 8397787.
- ↑ Chou, K. C.; Tomasselli, A. L.; Reardon, I. M.; Heinrikson, R. L. (Jan 1996). "Predicting HIV protease cleavage sites in proteins by a discriminant function method". Proteins 24 (1): 51–72. doi:10.1002/(SICI)1097-0134(199601)24:1<51::AID-PROT4>3.0.CO;2-R. PMID 8628733.
- ↑ Liang, G. Z.; Li, S. Z. (2007). "A new sequence representation as applied in better specificity elucidation for human immunodeficiency virus type 1 protease". Biopolymers 88 (3): 401–412. doi:10.1002/bip.20669. PMID 17206631.
- ↑ Shen, H.B.; Chou, K. C. (April 2008). "HIVcleave: a web-server for predicting HIV protease cleavage sites in proteins". Analytical Biochemistry 375 (2): 388–390. doi:10.1016/j.ab.2008.01.012. PMID 18249180.
- ↑ Gan, Y. R.; Huang, H.; Huang, Y. D.; Rao, C. M.; Zhao, Y.; Liu, J. S.; Wu, L.; Wei, D. Q. (April 2006). "Synthesis activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase". Peptides 27 (4): 622–625. doi:10.1016/j.peptides.2005.09.006. PMID 16242214.
- ↑ Du, Q. S.; Wang, S.; Wei, D. Q.; Sirois, S.; Chou, K. C. (2005). "Molecular modelling chemical modification for finding peptide inhibitor against SARS CoV Mpro.". Analytical Biochemistry 337: 262–270. doi:10.1016/j.ab.2004.10.003. PMID 15691506.
- ↑ Du, Q. S.; Sun, H.; Chou, K. C. (2007). "Inhibitor design for SARS coronavirus main protease based on "distorted key theory"". Medicinal Chemistry 3: 1–6. doi:10.2174/157340607779317616. PMID 17266617.

