Biology:Focal molography
Focal molography (“molography” in short) is a biophysical method for robust and sensitive detection of biomolecular interactions in a label-free manner. The new method enables biomolecular interaction analysis in complex biological samples without the use of additional fluorescent labels. Molography widens the analytic scope of biomolecular interaction analysis techniques in a broad range of applications, e.g. label-free trace analysis of a targeted molecule (called biomarker) in complex samples, such as blood sera, bioreactor fluid or cell culture media. Contrary to refractometric methods for label-free biomolecular interaction analysis, such as surface plasmon resonance (SPR) and reflectometric interference spectroscopy (RIfS), molography allows quantification of molecular interactions in living cells in real time.
Principle
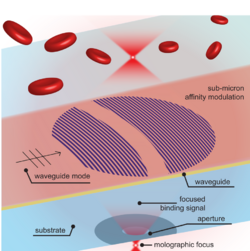
The working principle of molography is illustrated in Figure 1. Molography is based on diffraction of laser light at a special 2D nanopattern of molecular binding sites on the surface of a sensor chip, termed mologram. [1][2][3] A mologram is a coherent assembly of binding sites on a chip that form the blueprint of molecular hologram. [3][1][2] The hologram has the shape of a focusing diffractive lens and is illuminated by an evanescent wave. Evanescent waves occur when light is totally internally reflected at an interface of two dielectrics. [4][5] Biomolecules that bind to the mologram diffract laser light into a diffraction-limited focal spot in three dimensional space, the focal point of the mologram. [3][1][2] The intensity of the focused light correlates quadratically with the amount of bound molecules on the mologram and therefore also the number of biomolecular interactions that take place. [4][5]
In complex biological samples, the concentration of off-target molecules is usually significantly higher than the target molecules. Therefore, even in the absence of recognition elements the off-target molecules readily adsorb to the surface of the sensor. [3][1][2] However, this a random process and the off-target molecules do not bind to the ordered binding sites of the mologram. [3][1][2] Thus, scattering of off-target molecules is uniform in all spatial directions and therefore off-target molecules hardly contribute to the measured light intensity in the narrow solid angle of the focal spot. [3][1][2] In essence, focal molography forms a high-frequency spatial affinity lock-in that only measures the Fourier component of the refractive index distribution that corresponds to the binding modulation of the mologram. [6] Since the environmental noise (temperature gradients, buffer changes and nonspecific binding of off-target molecules) is inversely proportional to the spatial frequency and therefore situated mostly at "long" spatial periods most of the noise can be rejected. This property makes molography extremely robust and renders temperature stabilization or sensor equilibration obsolete. [7]
History
Christof Fattinger at Hoffmann-La-Roche in Basel already conceived the physical principles of focal molography in the 1990s when he investigated the bidiffractive grating-coupler. [1][8][9] Yet it only emerged in a scientific collaboration between ETH Zürich and the Roche Innovation Center Basel in Switzerland from 2014 to 2020.
Realization: A special photolithographic method enables the synthesis molograms
The synthesis of molograms on the sensor chip is realized with properly designed surface chemistry and reactive immersion lithography (RIL). [3][10] Through the RIL process the biomolecular recognition structure of the mologram on a light-sensitive non-fouling graft copolymer layer can be created by standard lithography techniques. [3][10][11] The copolymer layer is functionalized with photocleavable protection groups that upon illumination create reactive amines. [1][3][10] Subsequent surface chemistry steps enable an easy tailoring of recognition molecules specific to the desired analytical application. [10][12][13][14]
Applications of the molography method
The robustness of focal molography against environmental noise gives the method a platform character that enables many possible applications. [15] They range from the investigation of a specific biomolecular interaction in basic biological research to the diagnosis of a critical health condition in an emergency. [3][10]
The biological applications of molography categorize into five basic classes:
- Classical biomolecular interaction analysis (BIA),[16] i.e. the measurement of on-rates, off-rates and binding constants as demonstrated in,[10]
- Quantification of biomarkers in biological samples,[10]
- Profiling of low-affinity binders on mologram arrays,[13]
- Quantification of molecular interactions in living cells in real time,[12][14] and
- Discovery of unknown biomolecular interactions by means of binding assays.[1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Fattinger, Christof (2014-08-11). "Focal Molography: Coherent Microscopic Detection of Biomolecular Interaction". Physical Review X 4 (3): 031024. doi:10.1103/PhysRevX.4.031024. Bibcode: 2014PhRvX...4c1024F. https://link.aps.org/doi/10.1103/PhysRevX.4.031024.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Binnig, Gerd (2014-08-11). "Coherent Signal Picks Out Biomolecular Interactions" (in en). Physics 7: 84. doi:10.1103/Physics.7.84. ISSN 1943-2879. Bibcode: 2014PhyOJ...7...84B. https://link.aps.org/doi/10.1103/Physics.7.84.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 Fattinger, Christof, Frutiger, Andreas. "Focal Molography – an optical method for label-free detection of biomolecular interactions". https://www.sps.ch/en/artikel/progresses/focal-molography-an-optical-method-for-label-free-detection-of-biomolecular-interactions-77.
- ↑ 4.0 4.1 Blickenstorfer, Yves; Müller, Markus; Dreyfus, Roland; Reichmuth, Andreas Michael; Fattinger, Christof; Frutiger, Andreas (2021-03-09). "Quantitative Diffractometric Biosensing". Physical Review Applied 15 (3): 034023. doi:10.1103/PhysRevApplied.15.034023. Bibcode: 2021PhRvP..15c4023B. https://link.aps.org/doi/10.1103/PhysRevApplied.15.034023.
- ↑ 5.0 5.1 5.2 Frutiger, Andreas; Blickenstorfer, Yves; Bischof, Silvio; Forró, Csaba; Lauer, Matthias; Gatterdam, Volker; Fattinger, Christof; Vörös, János (2019-01-28). "Principles for Sensitive and Robust Biomolecular Interaction Analysis: The Limits of Detection and Resolution of Diffraction-Limited Focal Molography". Physical Review Applied 11 (1): 014056. doi:10.1103/PhysRevApplied.11.014056. Bibcode: 2019PhRvP..11a4056F. https://link.aps.org/doi/10.1103/PhysRevApplied.11.014056.
- ↑ Frutiger, Andreas; Fattinger, Christof; Vörös, János (January 2021). "Ultra-Stable Molecular Sensors by Sub-Micron Referencing and Why They Should Be Interrogated by Optical Diffraction—Part I. The Concept of a Spatial Affinity Lock-in Amplifier" (in en). Sensors 21 (2): 469. doi:10.3390/s21020469. PMID 33440783. Bibcode: 2021Senso..21..469F.
- ↑ Frutiger, Andreas; Gatterdam, Karl; Blickenstorfer, Yves; Reichmuth, Andreas Michael; Fattinger, Christof; Vörös, János (January 2021). "Ultra Stable Molecular Sensors by Submicron Referencing and Why They Should Be Interrogated by Optical Diffraction—Part II. Experimental Demonstration" (in en). Sensors 21 (1): 9. doi:10.3390/s21010009. PMID 33375003.
- ↑ Fattinger, Ch. (1993-03-29). "The bidiffractive grating coupler". Applied Physics Letters 62 (13): 1460–1462. doi:10.1063/1.108658. ISSN 0003-6951. Bibcode: 1993ApPhL..62.1460F. https://aip.scitation.org/doi/10.1063/1.108658.
- ↑ Spinke, J.; Oranth, N.; Fattinger, Ch.; Koller, H.; Mangold, C.; Voegelin, D. (1997-03-01). "The bidiffractive grating coupler: application to immunosensing" (in en). Sensors and Actuators B: Chemical 39 (1–3): 256–260. doi:10.1016/S0925-4005(97)80214-6. ISSN 0925-4005. https://www.sciencedirect.com/science/article/abs/pii/S0925400597802146.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 Gatterdam, Volker; Frutiger, Andreas; Stengele, Klaus-Peter; Heindl, Dieter; Lübbers, Thomas; Vörös, Janos; Fattinger, Christof (November 2017). "Focal molography is a new method for the in situ analysis of molecular interactions in biological samples" (in en). Nature Nanotechnology 12 (11): 1089–1095. doi:10.1038/nnano.2017.168. ISSN 1748-3395. PMID 28945239. Bibcode: 2017NatNa..12.1089G. https://www.nature.com/articles/nnano.2017.168.
- ↑ Frutiger, Andreas; Tschannen, Cla Duri; Blickenstorfer, Yves; Reichmuth, Andreas M.; Fattinger, Christof; Vörös, Janos (2018-12-01). "Image reversal reactive immersion lithography improves the detection limit of focal molography" (in EN). Optics Letters 43 (23): 5801–5804. doi:10.1364/OL.43.005801. ISSN 1539-4794. PMID 30499945. Bibcode: 2018OptL...43.5801F. https://www.osapublishing.org/ol/abstract.cfm?uri=ol-43-23-5801.
- ↑ 12.0 12.1 Reichmuth, Andreas Michael; Zimmermann, Mirjam; Wilhelm, Florian; Frutiger, Andreas; Blickenstorfer, Yves; Fattinger, Christof; Waldhoer, Maria; Vörös, János (2020-07-07). "Quantification of Molecular Interactions in Living Cells in Real Time using a Membrane Protein Nanopattern". Analytical Chemistry 92 (13): 8983–8991. doi:10.1021/acs.analchem.0c00987. ISSN 0003-2700. PMID 32524822. https://doi.org/10.1021/acs.analchem.0c00987.
- ↑ 13.0 13.1 Reichmuth, Andreas M.; Kübrich, Katharina; Blickenstorfer, Yves; Frutiger, Andreas; Momotenko, Dmitry; Gatterdam, Volker; Treindl, Fridolin; Fattinger, Christof et al. (2021-03-26). "Investigating Complex Samples with Molograms of Low-Affinity Binders". ACS Sensors 6 (3): 1067–1076. doi:10.1021/acssensors.0c02346. PMID 33629586. https://doi.org/10.1021/acssensors.0c02346.
- ↑ 14.0 14.1 Incaviglia, Ilaria; Frutiger, Andreas; Blickenstorfer, Yves; Treindl, Fridolin; Ammirati, Giulia; Lüchtefeld, Ines; Dreier, Birgit; Plückthun, Andreas et al. (2021-04-23). "An Approach for the Real-Time Quantification of Cytosolic Protein–Protein Interactions in Living Cells". ACS Sensors 6 (4): 1572–1582. doi:10.1021/acssensors.0c02480. PMID 33759497. https://doi.org/10.1021/acssensors.0c02480.
- ↑ Frutiger, Andreas (2021). Molecular Holograms - Design principles of robust biosensors at the example of focal molography (Doctoral Thesis thesis). ETH Zurich. doi:10.3929/ethz-b-000474270. hdl:20.500.11850/474270.
- ↑ Huber, Walter; Mueller, Francis (2006-11-01). "Biomolecular Interaction Analysis in Drug Discovery Using Surface Plasmon Resonance Technology" (in en). Current Pharmaceutical Design 12 (31): 3999–4021. doi:10.2174/138161206778743600. PMID 17100609. http://www.eurekaselect.com/openurl/content.php?genre=article&issn=1381-6128&volume=12&issue=31&spage=3999.
 |

