Biology:Actinomyces-1 RNA motif
From HandWiki
| Actinomyces-1 | |
|---|---|
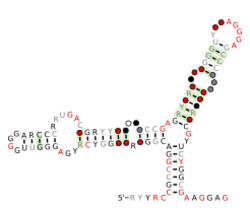 Consensus secondary structure and sequence conservation of Actinomyces-1 RNA | |
| Identifiers | |
| Symbol | Actinomyces-1 |
| Rfam | RF02928 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The Actinomyces-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] Actinomyces-1 motifs are found in the genus Actinomyces, in the phylum Actinomycetota. Actinomyces-1 RNAs likely function in trans as sRNAs. In terms of their secondary structure, Actinomyces-1 RNAs consist of a multistem junction with many conserved GA dinucleotides.
Actinomyces-1 RNAs contain a predicted kink turn.[2] This particular example of a kink turn was studied to better understand how kink turn structures relate to their sequences.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Research 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
- ↑ "Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules". Nucleic Acids Res 49 (10): 5916–5924. June 2021. doi:10.1093/nar/gkab333. PMID 33978763.
 |

