Biology:Avibirnavirus
| Avibirnavirus | |
|---|---|
| |
| Infectious bursal disease virus particle | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | incertae sedis |
| Family: | Birnaviridae |
| Genus: | Avibirnavirus |
| Species[1] | |
Avibirnavirus is a genus of viruses in family Birnaviridae.[2] There is a single species in this genus: Infectious bursal disease virus, which infects chickens and other fowl. It causes severe inflammation of the bursa of Fabricius, and causes considerable morbidity and mortality.[3][4]
History
Initially, the virus was discovered in 1957 in Gumboro, Delaware, United States .[5][6] Later, the disease was termed Gumboro disease. Since its discovery, the virus has been found to have a worldwide distribution.[6]
Structure
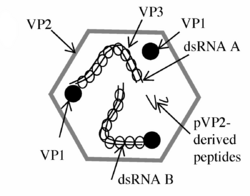
Avibirnaviruses are non-enveloped, and their single shelled icosahedral capsid exhibits a T=13 symmetry. The diameter of the capsid is around 70 nm.[3][4]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Avibirnavirus | Icosahedral | T=13 | Non-enveloped | Linear | Segmented |
Genome
The double-stranded RNA genome of Avibirnaviruses is linear and segmented into two segments (A and B).[7][8] The combined lengths of the two segments is around 6,000 nucleotides, almost entirely composed of reading-frames. The genome codes for 4-5 proteins depending on the strain.[3][4] The high degree of antigenic variation is due to a combination of high mutation rate and inter-strain homologous recombination within a hypervariable region that is located in the vp2 gene.[6][9]
At approximately 3,200 bp in length, segment A is the larger of the two segments. It contains two partially overlapping open reading frames (ORFs).[7][8] The first ORF in segment A encodes viral protein 5 (VP5).[7] The second ORF in segment A encodes a polyprotein, the pVP2-VP4-VP3 precursor, which is cleaved into three proteins .[7] Segment B encodes a single viral protein, VP1.[7][8]
Viral Proteins
VP1
Viral protein 1 is an RNA-dependent RNA polymerase, which cycles between the two segments and aids in the formation of ribonucleoprotein complexes with viral protein 3.[7]
VP2
Viral protein 2 is a major structural component of the virion.[7] The protein contains three domains: base, shell, and projection.[5] The projection domain consists of four loop structures that are exposed on the virion surface.[5] This protein is responsible for inducing a protective immune response, which neutralize monoclonal antibodies through binding to the project domain.[5] The viral protein in conjunction with VP5 acts as an apoptotic inducer, which causes affected cells to become cytotoxic.[9] Also, together with VP5, it inhibits cellular protein synthesis through activation of the protein kinase R pathway (PKR).[9] This activation leads to an increase in reactive oxygen species in the affected cells.[9]
VP3
Viral protein 3 is an immunogenic protein that interacts with VP1.[7] Its helps to regulate VP2 apoptosis by inhibiting phosphorylation of PKR and eukaryotic initiation factor 2 (eIF2).[9] Apoptotic regulation allows for the replication and release of the virus into other cells.[9]
VP4
Viral protein 4 is a serine protease, which catalyzes the hydrolysis of polyprotein pVP2-VP4-VP3 to release the viral proteins.[7] The serine protease that cleaves at ser-652 and lys-692.[7] This step is essential to replication due to release of the other proteins.[7] VP4 will additionally take over a glucocorticoid-induced leucine zipper protein (GILZ) that will allow for viral growth and block immune response of host cell.[7][9]
VP5
Viral protein 5 is an inducer of apoptosis for DF-1 cells.[10] It relies on inhibiting the voltage-dependent anion channel 2 (VDCA2) located on the mitochondria.[10] The inhibition will be aided by VP2, which will close the channel allowing for proliferation of the virus in host cells.[9][10] Also, VDCA2 will bind to RACK1 that will allow replication.[7]
Serotyping
The serotype and antigenic variations of the virus can only be distinguished between through virus-neutralization assays.[5] The project domains can be viewed to see the variation of the virus through amino acid substitutions.[5] The genome is typically highly conserved, however, the serotypes will vary the genome due to nucleotide changes.[8] There are two serotypes of avibirnavirus with one containing multiple classifications.[11] Serotype 1 is pathogenic to chickens especially at 3 to 6 weeks of age and seen to be more virulent in lighter breeds.[11][9] This serotype contains three main classifications: classical virulent IBDV, very virulent IBDV, and variant IBDV.[11][12] Classical virulent IBDV is the only serotype that has two subtypes: attenuated vaccine IBDV and classical virulent IBDV.[12] Although, these serotypes can have variance upon region.[5] Serotype 2 was identified in turkey.[5] This serotype has no pathogenic effect on chickens.[11]
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. The entry points in the host cell are through certain cell receptors.[11] One receptor that the virus will bind is surface immunoglobulin M, which specifically interacts with the light chains of the antibody.[9] Another receptor that can be bound is heat shock protein 90 (HSP90) on the surface of the DF-1 cell membrane.[9] Although, this receptor will bind either the virus or the VP2-viral sub particle.[9] Entry through the membrane has not been fully understood.[9] The penetration of the membrane is aided by the capsid PEP46.[9] The capid will have a peptide generated from the c-terminal of pVP2 that is released by VP4.[9] The peptide causes the membrane to become permeable by forming pores, which allow for PEP46 entry depending on calcium gradient.[9] If the inner calcium gradient is low, then it is thought to enter V-ATPase positive vesicles by endocytosis and uncoated for entry into the cytosol.[9] Also, it has been found VP2-α4β1 aids in spreading the virus in the cell as well through macropinocytosis.[9] This will allow for the virus to be moved to the early endosomes that is accomplished in the cell through Rab5.[9] Replication follows the double-stranded RNA virus replication model. Double-stranded rna virus transcription is the method of transcription. Young chickens and other fowl serve as the natural host. Transmission routes are contamination.[3][4]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Avibirnavirus | Birds | None | Cell receptor endocytosis | Budding | Cytoplasm | Cytoplasm | Contact |
References
- ↑ "Virus Taxonomy: 2018b Release" (in en). March 2019. https://ictv.global/taxonomy. Retrieved 8 November 2019.
- ↑ Delmas, B; Attoui, H; Ghosh, S; Malik, YS; Mundt, E; Vakharia, VN; Ictv Report, Consortium (January 2019). "ICTV virus taxonomy profile: Birnaviridae.". The Journal of General Virology 100 (1): 5–6. doi:10.1099/jgv.0.001185. PMID 30484762.
- ↑ 3.0 3.1 3.2 3.3 "ICTV Report Birnaviridae". http://www.ictv.global/report/birnaviridae.
- ↑ 4.0 4.1 4.2 4.3 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/572.html. Retrieved 15 June 2015.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Jackwood, Daral J.; Schat, Karel A.; Michel, Linda O.; Wit, Sjaak de (2018-11-02). "A proposed nomenclature for infectious bursal disease virus isolates". Avian Pathology 47 (6): 576–584. doi:10.1080/03079457.2018.1506092. ISSN 0307-9457. PMID 30086652.
- ↑ 6.0 6.1 6.2 He, Xiumiao; Wang, Weiwei; Chen, Guo; Jiao, Pengtao; Ji, Zhonghua; Yang, Lin; Wei, Ping (2019-12-01). "Serological study reveal different antigenic IBDV strains prevalent in southern China during the years 2000–2017 and also the antigenic differences between the field strains and the commonly used vaccine strains" (in en). Veterinary Microbiology 239: 108458. doi:10.1016/j.vetmic.2019.108458. ISSN 0378-1135. PMID 31767074. http://www.sciencedirect.com/science/article/pii/S0378113519308351.
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 7.11 7.12 Wang, Nian; Zhang, Lizhou; Chen, Yuming; Lu, Zhen; Gao, Li; Wang, Yongqiang; Gao, Yulong; Gao, Honglei et al. (2015). "Cyclophilin A Interacts with Viral VP4 and Inhibits the Replication of Infectious Bursal Disease Virus". BioMed Research International 2015: 719454. doi:10.1155/2015/719454. ISSN 2314-6133. PMID 26090438.
- ↑ 8.0 8.1 8.2 8.3 Kibenge, F. S. B.; Nagarajan, M. M.; Qian, B. (June 1996). "Determination of the 5′ and 3′ terminal noncoding sequences of the bi-segmented genome of the avibirnavirus infectious bursal disease virus". Archives of Virology 141 (6): 1133–1141. doi:10.1007/bf01718616. ISSN 0304-8608. PMID 8712930.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 9.14 9.15 9.16 9.17 9.18 Qin, Yao; Zheng, Shijun J. (2017-01-14). "Infectious Bursal Disease Virus-Host Interactions: Multifunctional Viral Proteins that Perform Multiple and Differing Jobs". International Journal of Molecular Sciences 18 (1): 161. doi:10.3390/ijms18010161. ISSN 1422-0067. PMID 28098808.
- ↑ 10.0 10.1 10.2 (in en) Avibirnavirus—Advances in Research and Application: 2012 Edition: ScholarlyPaper. ScholarlyEditions. 2012-12-26. ISBN 978-1-4816-2427-5. https://books.google.com/books?id=ih_ZsAePZ64C&q=Avibirnavirus&pg=PT4.
- ↑ 11.0 11.1 11.2 11.3 11.4 Rekha, Kaliyaperumal; Sivasubramanian, Chandran; Chung, Ill-Min; Thiruvengadam, Muthu (2014). "Growth and Replication of Infectious Bursal Disease Virus in the DF-1 Cell Line and Chicken Embryo Fibroblasts". BioMed Research International 2014: 494835. doi:10.1155/2014/494835. ISSN 2314-6133. PMID 24949455.
- ↑ 12.0 12.1 Tomás, Gonzalo; Hernández, Martín; Marandino, Ana; Techera, Claudia; Grecco, Sofia; Hernández, Diego; Banda, Alejandro; Panzera, Yanina et al. (2017-03-04). "Development of an RT-qPCR assay for the specific detection of a distinct genetic lineage of the infectious bursal disease virus". Avian Pathology 46 (2): 150–156. doi:10.1080/03079457.2016.1228827. ISSN 0307-9457. PMID 27924642.
External links
Wikidata ☰ Q790863 entry
 |

