Biology:C20orf96
C20orf96 (Chromosome 20 open reading frame 96) is a protein-coding gene in humans.[1] It codes for an unknown protein known as uncharacterized protein C20orf96, predicted to be a nuclear protein. The function and biological processes of the gene is not well understood by the scientific community yet.
Gene aliases
C20orf96 is also known by the alias DJ1103G7.2. Orthologs found in other organisms are known as C20orf96 homolog isoform X [Species].
Gene
Location
C20orf96 is located on the short arm of chromosome 20 at 20p13, base pairs 270,863 to 290,778 on the complementary strand. It is a member of the DUF4618 superfamily, with the DUF position being from amino acid 104 to 363.[1] Neighboring genes are DEFB129, DEFB132, ZCCHC3, and NRSN2-AS1.
Expression
Aceview states that the gene is expressed 2.2 times the amount an average gene is expressed, and the sequence has been seen in the brain, testis, uterus, kidney, thymus, breast, kidney tumor, and 84 other places.[2] C20orf96 has been found to bind to transcription factor binding sites AREB6, GATA-1, GATA-2, GATA-3, ATF6, c-Myc, Max, CHOP-10, AMLa, and C/EBPalpha.[3] TYW5 and ALOXE3 are two proteins that interact with C20orf96. TYW5 is a protein coding gene and also incorporates tRNA modification in the nucleus.[4] ALOXE3 is also a protein coding gene, more focusing on Prostaglandin 2 biosynthesis.[5]
mRNA
Alternative splicing
C20orf96 can be split up into five different variants. All variants code for similar proteins. The variant used for this article was Variant 1. The splicing is shown below.
| Exon | Exon Size (bp) |
|---|---|
| 1 | 18 |
| 2 | 48 |
| 3 | 117 |
| 4 | 117 |
| 5 | 159 |
| 6 | 99 |
| 7 | 156 |
| 8 | 102 |
| 9 | 171 |
| 10 | 117 |
| 11 | 60 |
Protein
General properties
The protein made by C20orf96 is 363 amino acids in length. The predicted molecular weight is 42.9 kdal, and the isoelectric point is 8.99.[6][7]
Composition
The ratios of this protein are similar to others. The unusual amounts of amino acids are found as alanine, glutamic acid, glycine, and glutamine. Glutamic acid and glutamine both have a slightly larger amount than normal, alanine is slightly below normal, and glycine is low compared to other proteins.
A- : 13( 3.6%) |
C : 4( 1.1%) |
D : 15( 4.1%) |
E+ : 38(10.5%) |
F : 10( 2.8%) |
|---|---|---|---|---|
G--: 4( 1.1%) |
H : 9( 2.5%) |
I : 17( 4.7%) |
K : 34( 9.4%) |
L : 40(11.0%) |
M : 13( 3.6%) |
N : 10( 2.8%) |
P : 19( 5.2%) |
Q+ : 35( 9.6%) |
R : 24( 6.6%) |
S : 25( 6.9%) |
T : 21( 5.8%) |
V : 22( 6.1%) |
W : 4( 1.1%) |
Y : 6( 1.7%) |
C20orf96 has a neutral charge, with no additional positive, negative, or mixed charge clusters.
Post-translational modification
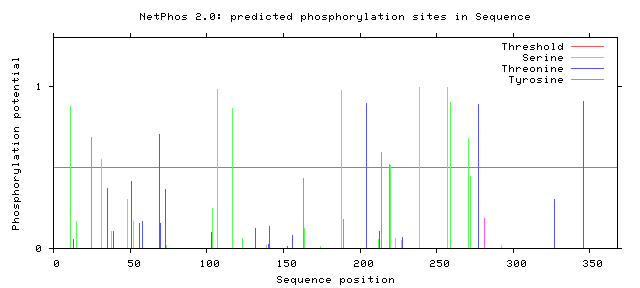
NetPhos predicts 10 serine sites, 4 threonine sites, and 3 tyrosine sites over the 363 amino acids.[8] No signal peptides were detected. There were also no transmembrane sequences.
Secondary structure
Most of the structure is made up of alpha-helices, with short coils on both ends. At the end of the sequence, there is also a small, four amino acid beta-strand.
Tertiary structure
There are no known crystal structures for this protein. The tertiary structure is mainly composed of coiled-coil regions.
Homology
C20orf96 is found in many eukaryotes. Orthologs have been found in most organisms in the kingdom animalia, with the lineage going back to the phylum Chordata. No paralogs for C20orf96 have been found.
Mutations
The most prevalent mutations of C20orf96 in humans are M1V, Q6K, T13I, T13S, M94I, S117G, S117T, S117N, L142V, D159N, M215T, D238E, R251C, R251G, Q263E, R279C, E304G, I305F, M313I, R328W, R342I, and L353V. The format these mutations are given in are the amino acid that is supposed to be there, the amino acid number, and then the mutation.
Connection to diseases
C20orf96 has been linked to Moyamoya Disease, increasing the odds of contracting the disease by 4.5 times.[9] It is also associated with lymph node metastasis in colorectal cancer.[10] A study found that C20orf96 has shown positive selection for containing a hub gene, which is a gene that is ranked in the top 20 for connectivity.[11]
References
- ↑ 1.0 1.1 "C20orf96 chromosome 20 open reading frame 96 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/140680.
- ↑ Thierry-Mieg, Danielle; Thierry-Mieg, Jean. "AceView: Gene:C20orf96, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView.". https://www.ncbi.nlm.nih.gov/ieb/research/acembly/av.cgi?db=human&term=c20orf96&submit=Go.
- ↑ "www.genecards.org/cgi-bin/carddisp.pl?gene=C20orf96". https://www.genecards.org/cgi-bin/carddisp.pl?gene=C20orf96.
- ↑ "www.genecards.org/cgi-bin/carddisp.pl?gene=tyw5". https://www.genecards.org/cgi-bin/carddisp.pl?gene=tyw5.
- ↑ "www.genecards.org/cgi-bin/carddisp.pl?gene=ALOXE3&keywords=aloxe3". https://www.genecards.org/cgi-bin/carddisp.pl?gene=ALOXE3&keywords=aloxe3.
- ↑ "SAPS". SDSC Biology Workbench. Retrieved 2016-05-08.
- ↑ "IP". SDSC Biology Workbench. Retrieved 2016-05-08.
- ↑ "NetPhos 2.0 Server". www.cbs.dtu.dk. Retrieved 2016-05-08.
- ↑ Sigdel, T., Shoemaker, L., Chen, R., Li, L., Butte, A., Sarwal, M. and Steinberg, G. (2013). Immune response profiling identifies autoantibodies specific to Moyamoya patients. Orphanet Journal of Rare Diseases, 8(1), p.45.
- ↑ Sasaki, K., Oga, A., Furuya, T., Chochi, Y., Ito, H., Adachi, J., Uchiyama, T., Kawauchi, S. and Nakao, M. (2011). DNA copy number aberrations associated with the clinicopathological features of colorectal cancers: Identification of genomic biomarkers by array-based comparative genomic hybridization. Oncology Reports, 25, pp.1603-1611.
- ↑ Konopka, G., Friedrich, T., Davis-Turak, J., Winden, K., Oldham, M., Gao, F., Chen, L., Wang, G., Luo, R., Preuss, T. and Geschwind, D. (2012). Human-Specific Transcriptional Networks in the Brain. Neuron, 75(4), pp.601-617.
 |