Biology:Cladistics
Cladistics (/kləˈdɪstɪks/; from grc κλάδος (kládos) 'branch')[1] is an approach to biological classification in which organisms are categorized in groups ("clades") based on hypotheses of most recent common ancestry. The evidence for hypothesized relationships is typically shared derived characteristics (synapomorphies) that are not present in more distant groups and ancestors. However, from an empirical perspective, common ancestors are inferences based on a cladistic hypothesis of relationships of taxa whose character states can be observed. Theoretically, a last common ancestor and all its descendants constitute a (minimal) clade. Importantly, all descendants stay in their overarching ancestral clade. For example, if the terms worms or fishes were used within a strict cladistic framework, these terms would include humans. Many of these terms are normally used paraphyletically, outside of cladistics, e.g. as a 'grade', which are fruitless to precisely delineate, especially when including extinct species. Radiation results in the generation of new subclades by bifurcation, but in practice sexual hybridization may blur very closely related groupings.[2][3][4]
As a hypothesis, a clade can be rejected only if some groupings were explicitly excluded. It may then be found that the excluded group did actually descend from the last common ancestor of the group, and thus emerged within the group. ("Evolved from" is misleading, because in cladistics all descendants stay in the ancestral group). To keep only valid clades, upon finding that the group is paraphyletic this way, either such excluded groups should be granted to the clade, or the group should be abolished.[5]
Branches down to the divergence to the next significant (e.g. extant) sister are considered stem-groupings of the clade, but in principle each level stands on its own, to be assigned a unique name. For a fully bifurcated tree, adding a group to a tree also adds an additional (named) clade, and a new level on that branch. Specifically, also extinct groups are always put on a side-branch, not distinguishing whether an actual ancestor of other groupings was found.
The techniques and nomenclature of cladistics have been applied to disciplines other than biology. (See phylogenetic nomenclature.)
Cladistics findings are posing a difficulty for taxonomy, where the rank and (genus-)naming of established groupings may turn out to be inconsistent.
Cladistics is now the most commonly used method to classify organisms.[6]
History



The original methods used in cladistic analysis and the school of taxonomy derived from the work of the German entomologist Willi Hennig, who referred to it as phylogenetic systematics (also the title of his 1966 book); but the terms "cladistics" and "clade" were popularized by other researchers. Cladistics in the original sense refers to a particular set of methods used in phylogenetic analysis, although it is now sometimes used to refer to the whole field.[7]
What is now called the cladistic method appeared as early as 1901 with a work by Peter Chalmers Mitchell for birds[8][9] and subsequently by Robert John Tillyard (for insects) in 1921,[10] and W. Zimmermann (for plants) in 1943.[11] The term "clade" was introduced in 1958 by Julian Huxley after having been coined by Lucien Cuénot in 1940,[12] "cladogenesis" in 1958,[13] "cladistic" by Arthur Cain and Harrison in 1960,[14] "cladist" (for an adherent of Hennig's school) by Ernst Mayr in 1965,[15] and "cladistics" in 1966.[13] Hennig referred to his own approach as "phylogenetic systematics". From the time of his original formulation until the end of the 1970s, cladistics competed as an analytical and philosophical approach to systematics with phenetics and so-called evolutionary taxonomy. Phenetics was championed at this time by the numerical taxonomists Peter Sneath and Robert Sokal, and evolutionary taxonomy by Ernst Mayr.Lua error: Internal error: The interpreter exited with status 1.
Originally conceived, if only in essence, by Willi Hennig in a book published in 1950, cladistics did not flourish until its translation into English in 1966 (Lewin 1997). Today, cladistics is the most popular method for inferring phylogenetic trees from morphological data.
In the 1990s, the development of effective polymerase chain reaction techniques allowed the application of cladistic methods to biochemical and molecular genetic traits of organisms, vastly expanding the amount of data available for phylogenetics. At the same time, cladistics rapidly became popular in evolutionary biology, because computers made it possible to process large quantities of data about organisms and their characteristics.
Methodology
Lua error: Internal error: The interpreter exited with status 1.
The cladistic method interprets each shared character state transformation as a potential piece of evidence for grouping. Synapomorphies (shared, derived character states) are viewed as evidence of grouping, while symplesiomorphies (shared ancestral character states) are not. The outcome of a cladistic analysis is a cladogram – a tree-shaped diagram (dendrogram)[16] that is interpreted to represent the best hypothesis of phylogenetic relationships. Although traditionally such cladograms were generated largely on the basis of morphological characters and originally calculated by hand, genetic sequencing data and computational phylogenetics are now commonly used in phylogenetic analyses, and the parsimony criterion has been abandoned by many phylogeneticists in favor of more "sophisticated" but less parsimonious evolutionary models of character state transformation. Cladists contend that these models are unjustified because there is no evidence that they recover more "true" or "correct" results from actual empirical data sets [17]
Every cladogram is based on a particular dataset analyzed with a particular method. Datasets are tables consisting of molecular, morphological, ethological[18] and/or other characters and a list of operational taxonomic units (OTUs), which may be genes, individuals, populations, species, or larger taxa that are presumed to be monophyletic and therefore to form, all together, one large clade; phylogenetic analysis infers the branching pattern within that clade. Different datasets and different methods, not to mention violations of the mentioned assumptions, often result in different cladograms. Only scientific investigation can show which is more likely to be correct.
Until recently, for example, cladograms like the following have generally been accepted as accurate representations of the ancestral relations among turtles, lizards, crocodilians, and birds:[19] Lua error: Internal error: The interpreter exited with status 1.
If this phylogenetic hypothesis is correct, then the last common ancestor of turtles and birds, at the branch near the ▼ lived earlier than the last common ancestor of lizards and birds, near the ♦. Most molecular evidence, however, produces cladograms more like this:[20] Lua error: Internal error: The interpreter exited with status 1.
If this is accurate, then the last common ancestor of turtles and birds lived later than the last common ancestor of lizards and birds. Since the cladograms show two mutually exclusive hypotheses to describe the evolutionary history, at most one of them is correct.
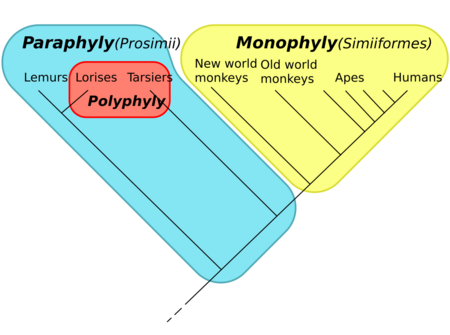
The cladogram to the right represents the current universally accepted hypothesis that all primates, including strepsirrhines like the lemurs and lorises, had a common ancestor all of whose descendants are or were primates, and so form a clade; the name Primates is therefore recognized for this clade. Within the primates, all anthropoids (monkeys, apes, and humans) are hypothesized to have had a common ancestor all of whose descendants are or were anthropoids, so they form the clade called Anthropoidea. The "prosimians", on the other hand, form a paraphyletic taxon. The name Prosimii is not used in phylogenetic nomenclature, which names only clades; the "prosimians" are instead divided between the clades Strepsirhini and Haplorhini, where the latter contains Tarsiiformes and Anthropoidea.
Lemurs and tarsiers may have looked closely related to humans, in the sense of being close on the evolutionary tree to humans. However, from the perspective of a tarsier, humans and lemurs would have looked close, in the exact same sense. Cladistics forces a neutral perspective, treating all branches (extant or extinct) in the same manner. It also forces one to try to make statements, and honestly take into account findings, about the exact historic relationships between the groups.
Terminology for character states
The following terms, coined by Hennig, are used to identify shared or distinct character states among groups:[21][22][23]
- Lua error: Internal error: The interpreter exited with status 1. A plesiomorphy ("close form") or ancestral state is a character state that a taxon has retained from its ancestors. When two or more taxa that are not nested within each other share a plesiomorphy, it is a symplesiomorphy (from syn-, "together"). Symplesiomorphies do not mean that the taxa that exhibit that character state are necessarily closely related. For example, Reptilia is traditionally characterized by (among other things) being cold-blooded (i.e., not maintaining a constant high body temperature), whereas birds are warm-blooded. Since cold-bloodedness is a plesiomorphy, inherited from the common ancestor of traditional reptiles and birds, and thus a symplesiomorphy of turtles, snakes and crocodiles (among others), it does not mean that turtles, snakes and crocodiles form a clade that excludes the birds.
- Lua error: Internal error: The interpreter exited with status 1.Lua error: Internal error: The interpreter exited with status 1. An apomorphy ("separate form") or derived state is an innovation. It can thus be used to diagnose a clade – or even to help define a clade name in phylogenetic nomenclature. Features that are derived in individual taxa (a single species or a group that is represented by a single terminal in a given phylogenetic analysis) are called autapomorphies (from auto-, "self"). Autapomorphies express nothing about relationships among groups; clades are identified (or defined) by synapomorphies (from syn-, "together"). For example, the possession of digits that are homologous with those of Homo sapiens is a synapomorphy within the vertebrates. The tetrapods can be singled out as consisting of the first vertebrate with such digits homologous to those of Homo sapiens together with all descendants of this vertebrate (an apomorphy-based phylogenetic definition).[24] Importantly, snakes and other tetrapods that do not have digits are nonetheless tetrapods: other characters, such as amniotic eggs and diapsid skulls, indicate that they descended from ancestors that possessed digits which are homologous with ours.
- A character state is homoplastic or "an instance of homoplasy" if it is shared by two or more organisms but is absent from their common ancestor or from a later ancestor in the lineage leading to one of the organisms. It is therefore inferred to have evolved by convergence or reversal. Both mammals and birds are able to maintain a high constant body temperature (i.e., they are warm-blooded). However, the accepted cladogram explaining their significant features indicates that their common ancestor is in a group lacking this character state, so the state must have evolved independently in the two clades. Warm-bloodedness is separately a synapomorphy of mammals (or a larger clade) and of birds (or a larger clade), but it is not a synapomorphy of any group including both these clades. Hennig's Auxiliary Principle[25] states that shared character states should be considered evidence of grouping unless they are contradicted by the weight of other evidence; thus, homoplasy of some feature among members of a group may only be inferred after a phylogenetic hypothesis for that group has been established.
The terms plesiomorphy and apomorphy are relative; their application depends on the position of a group within a tree. For example, when trying to decide whether the tetrapods form a clade, an important question is whether having four limbs is a synapomorphy of the earliest taxa to be included within Tetrapoda: did all the earliest members of the Tetrapoda inherit four limbs from a common ancestor, whereas all other vertebrates did not, or at least not homologously? By contrast, for a group within the tetrapods, such as birds, having four limbs is a plesiomorphy. Using these two terms allows a greater precision in the discussion of homology, in particular allowing clear expression of the hierarchical relationships among different homologous features.
It can be difficult to decide whether a character state is in fact the same and thus can be classified as a synapomorphy, which may identify a monophyletic group, or whether it only appears to be the same and is thus a homoplasy, which cannot identify such a group. There is a danger of circular reasoning: assumptions about the shape of a phylogenetic tree are used to justify decisions about character states, which are then used as evidence for the shape of the tree.[26] Phylogenetics uses various forms of parsimony to decide such questions; the conclusions reached often depend on the dataset and the methods. Such is the nature of empirical science, and for this reason, most cladists refer to their cladograms as hypotheses of relationship. Cladograms that are supported by a large number and variety of different kinds of characters are viewed as more robust than those based on more limited evidence.[27]
Terminology for taxa
Mono-, para- and polyphyletic taxa can be understood based on the shape of the tree (as done above), as well as based on their character states.[22][23][28] These are compared in the table below.
| Term | Node-based definition | Character-based definition |
|---|---|---|
| Holophyly, Monophyly | A clade, a monophyletic taxon, is a taxon that consists of the last common ancestor and all its descendants.[29] | A clade is characterized by one or more apomorphies: derived character states present in the first member of the taxon, inherited by its descendants (unless secondarily lost), and not inherited by any other taxa. |
| Paraphyly | A paraphyletic assemblage is one that is constructed by taking a clade and removing one or more smaller clades.[30] (Removing one clade produces a singly paraphyletic assemblage, removing two produces a doubly paraphylectic assemblage, and so on.)[31] | A paraphyletic assemblage is characterized by one or more plesiomorphies: character states inherited from ancestors but not present in all of their descendants. As a consequence, a paraphyletic assemblage is truncated, in that it excludes one or more clades from an otherwise monophyletic taxon. An alternative name is evolutionary grade, referring to an ancestral character state within the group. While paraphyletic assemblages are popular among paleontologists and evolutionary taxonomists, cladists do not recognize paraphyletic assemblages as having any formal information content – they are merely parts of clades. |
| Polyphyly | A polyphyletic assemblage is one which is neither monophyletic nor paraphyletic. | A polyphyletic assemblage is characterized by one or more homoplasies: character states which have converged or reverted so as to be the same but which have not been inherited from a common ancestor. No systematist recognizes polyphyletic assemblages as taxonomically meaningful entities, although ecologists sometimes consider them meaningful labels for functional participants in ecological communities (e. g., primary producers, detritivores, etc.). |
Criticism
Cladistics, either generally or in specific applications, has been criticized from its beginnings. Decisions as to whether particular character states are homologous, a precondition of their being synapomorphies, have been challenged as involving circular reasoning and subjective judgements.[32] Of course, the potential unreliability of evidence is a problem for any systematic method, or for that matter, for any empirical scientific endeavor at all.[33][34]
Transformed cladistics arose in the late 1970s [35] in an attempt to resolve some of these problems by removing a priori assumptions about phylogeny from cladistic analysis, but it has remained unpopular.[36]
Issues
Ancestors
The cladistic method does not identify fossil species as actual ancestors of a clade.[37] Instead, fossil taxa are identified as belonging to separate extinct branches. While a fossil species could be the actual ancestor of a clade, there is no way to know that. Therefore, a more conservative hypothesis is that the fossil taxon is related to other fossil and extant taxa, as implied by the pattern of shared apomorphic features.[38]
Extinction status
An otherwise extinct group with any extant descendants, is not considered (literally) extinct,[39] and for instance does not have a date of extinction.
Hybridization, interbreeding
Anything having to do with biology and sex is complicated and messy, and cladistics is no exception.[40] Many species reproduce sexually, and are capable of interbreeding for millions of years. Worse, during such a period, many branches may have radiated, and it may take hundreds of millions of years for them to have whittled down to just two.[41] Only then one can theoretically assign proper last common ancestors of groupings which do not inadvertently include earlier branches.[42] The process of true cladistic bifurcation can thus take a much more extended time than one is usually aware of.[43] In practice, for recent radiations, cladistically guided findings only give a coarse impression of the complexity. A more detailed account will give details about fractions of introgressions between groupings, and even geographic variations thereof. This has been used as an argument for the use of paraphyletic groupings,[42] but typically other reasons are quoted.
Horizontal gene transfer
Horizontal gene transfer is the mobility of genetic info between different organisms that can have immediate or delayed effects for the reciprocal host.[44] There are several processes in nature which can cause horizontal gene transfer. This does typically not directly interfere with ancestry of the organism, but can complicate the determination of that ancestry. On another level, one can map the horizontal gene transfer processes, by determining the phylogeny of the individual genes using cladistics.
Naming stability
If there is unclarity in mutual relationships, there are a lot of possible trees. Assigning names to each possible clade may not be prudent. Furthermore, established names are discarded in cladistics, or alternatively carry connotations which may no longer hold, such as when additional groups are found to have emerged in them.[45] Naming changes are the direct result of changes in the recognition of mutual relationships, which often is still in flux, especially for extinct species. Hanging on to older naming and/or connotations is counter-productive, as they typically do not reflect actual mutual relationships precisely at all. E.g. Archaea, Asgard archaea, protists, slime molds, worms, invertebrata, fishes, reptilia, monkeys, Ardipithecus, Australopithecus, Homo erectus all contain Homo sapiens cladistically, in their sensu lato meaning. For originally extinct stem groups, sensu lato generally means generously keeping previously included groups, which then may come to include even living species. A pruned sensu stricto meaning is often adopted instead, but the group would need to be restricted to a single branch on the stem. Other branches then get their own name and level. This is commensurate to the fact that more senior stem branches are in fact closer related to the resulting group than the more basal stem branches; that those stem branches only may have lived for a short time does not affect that assessment in cladistics.
In disciplines other than biology
The comparisons used to acquire data on which cladograms can be based are not limited to the field of biology.[46] Any group of individuals or classes that are hypothesized to have a common ancestor, and to which a set of common characteristics may or may not apply, can be compared pairwise. Cladograms can be used to depict the hypothetical descent relationships within groups of items in many different academic realms. The only requirement is that the items have characteristics that can be identified and measured.
Anthropology and archaeology:[47] Cladistic methods have been used to reconstruct the development of cultures or artifacts using groups of cultural traits or artifact features.
Comparative mythology and folktale use cladistic methods to reconstruct the protoversion of many myths. Mythological phylogenies constructed with mythemes clearly support low horizontal transmissions (borrowings), historical (sometimes Palaeolithic) diffusions and punctuated evolution.[48] They also are a powerful way to test hypotheses about cross-cultural relationships among folktales.[49][50]
Literature: Cladistic methods have been used in the classification of the surviving manuscripts of the Canterbury Tales,[51] and the manuscripts of the Sanskrit Charaka Samhita.[52]
Historical linguistics:[53] Cladistic methods have been used to reconstruct the phylogeny of languages using linguistic features. This is similar to the traditional comparative method of historical linguistics, but is more explicit in its use of parsimony and allows much faster analysis of large datasets (computational phylogenetics).
Textual criticism or stemmatics:[52][54] Cladistic methods have been used to reconstruct the phylogeny of manuscripts of the same work (and reconstruct the lost original) using distinctive copying errors as apomorphies. This differs from traditional historical-comparative linguistics in enabling the editor to evaluate and place in genetic relationship large groups of manuscripts with large numbers of variants that would be impossible to handle manually. It also enables parsimony analysis of contaminated traditions of transmission that would be impossible to evaluate manually in a reasonable period of time.
Astrophysics[55] infers the history of relationships between galaxies to create branching diagram hypotheses of galaxy diversification.
See also
Notes and references
- ↑ Template:Cite OEtymD
- ↑ Columbia Encyclopedia
- ↑ "Introduction to Cladistics". Ucmp.berkeley.edu. http://www.ucmp.berkeley.edu/clad/clad1.html.
- ↑ Oxford Dictionary of English{{full citation needed|date=April 2019} tionaryLua error: Internal error: The interpreter exited with status 1.
- ↑ Hickman, Cleveland P. Jr. (2014). Integrated principles of zoology (Sixteenth ed.). New York. ISBN 978-0-07-352421-4. OCLC 846846729. https://www.worldcat.org/oclc/846846729.
- ↑ "The Need for Cladistics". http://www.ucmp.berkeley.edu/clad/clad5.html.
- ↑ Brinkman & Leipe 2001, p. 323
- ↑ Schuh, Randall. 2000. Biological Systematics: Principles and Applications, p.7 (citing Nelson and Platnick, 1981). Cornell University Press (books.google)
- ↑ Folinsbee, Kaila et al. 2007. 5 Quantitative Approaches to Phylogenetics, p. 172. Rev. Mex. Div. 225-52 (kfolinsb.public.iastate.edu)
- ↑ Craw, RC (1992). "Margins of cladistics: Identity, differences and place in the emergence of phylogenetic systematics". in Griffiths, PE. Trees of life: Essays in the philosophy of biology. Dordrecht: Kluwer Academic. pp. 65–107. ISBN 978-94-015-8038-0.
- ↑ Schuh, Randall. 2000. Biological Systematics: Principles and Applications, p.7. Cornell U. Press
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ 13.0 13.1 Webster's 9th New Collegiate DictionaryLua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Rindal, Eirik; Brower, Andrew V. Z. (2011), "Do model-based phylogenetic analyses perform better than parsimony? A test with empirical data", Cladistics 27 (3): 331–334, doi:10.1111/j.1096-0031.2010.00342.x, PMID 34875779
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Benton, Michael J. (2005), Vertebrate Palaeontology, Blackwell, pp. 214, 233, ISBN 978-0-632-05637-8
- ↑ Lyson, Tyler; Gilbert, Scott F. (March–April 2009), "Turtles all the way down: loggerheads at the root of the chelonian tree", Evolution & Development 11 (2): 133–135, doi:10.1111/j.1525-142X.2009.00325.x, PMID 19245543, http://www.mrfdigs.com/publications/2009_lyson-gilbert-loggerheads.pdf
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ 22.0 22.1 Lua error: Internal error: The interpreter exited with status 1.
- ↑ 23.0 23.1 Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.: "Synapomorphies are invoked to defend the hypothesis; the hypothesis is invoked to defend the synapomorphies."
- ↑ Brower, AVZ and Schuh, RT. 2021. Biological Systematics: Principles and Applications (3rd edn.). Cornell University Press, Ithaca nY
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Podani, János (2010-08-01). "Monophyly and paraphyly: A discourse without end?" (in en). Taxon 59 (4): 1011–1015. doi:10.1002/tax.594002.
- ↑ Many sources give a verbal definition of 'paraphyletic' that does not require the missing groups to be monophyletic. However, when diagrams are presented representing paraphyletic groups, these invariably show the missing groups as monophyletic. See e.g.Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Oreskes, Naomi, Kristin Shrader-Frechette, and Kenneth Belitz. "Verification, validation, and confirmation of numerical models in the earth sciences." Science 263, no. 5147 (1994): 641-646.
- ↑ Nils Møller Anderson, 2001 The impact of W. Hennig’s “phylogenetic systematics” on contemporary entomology Eur. J.Entomol. 98: 133-150 online
- ↑ Platnick, Norman I. "Philosophy and the transformation of cladistics." Systematic Zoology 28, no. 4 (1979): 537–546.
- ↑ Brower, Andrew VZ. "Fifty shades of cladism." Biology & Philosophy 33, no. 1-2 (2018): 8.
- ↑ Krell, Frank-T; Cranston, Peter S. (2004). "Which side of the tree is more basal?: Editorial" (in en). Systematic Entomology 29 (3): 279–281. doi:10.1111/j.0307-6970.2004.00262.x.
- ↑ Patterson, Colin. "Significance of fossils in determining evolutionary relationships." Annual Review of Ecology and Systematics 12, no. 1 (1981): 195–223.
- ↑ Ross, Robert M.; Allmon, Warren D. (1990-12-18) (in en). Causes of Evolution: A Paleontological Perspective. University of Chicago Press. pp. 133. ISBN 978-0-226-72824-7. https://books.google.com/books?id=gw3vHelHbusC&dq=paraphyly+can+not+be+precisely+extinction&pg=PA133.
- ↑ "Introduction to Cladistics". https://ucmp.berkeley.edu/clad/clad1.html.
- ↑ Harrison, Richard G.; Larson, Erica L. (2014-01-01). "Hybridization, Introgression, and the Nature of Species Boundaries". Journal of Heredity 105 (S1): 795–809. doi:10.1093/jhered/esu033. ISSN 0022-1503. PMID 25149255.
- ↑ 42.0 42.1 Hörandl, Elvira; Stuessy, Tod F. (2010). "Paraphyletic groups as natural units of biological classification". Taxon 59 (6): 1641–1653. doi:10.1002/tax.596001. ISSN 0040-0262. https://www.jstor.org/stable/41059863.
- ↑ Mehta, Rohan S.; Rosenberg, Noah A. (2019-10-01). "The probability of reciprocal monophyly of gene lineages in three and four species" (in en). Theoretical Population Biology 129: 133–147. doi:10.1016/j.tpb.2018.04.004. PMID 29729946.
- ↑ Emamalipour, Melissa; Seidi, Khaled; Zununi Vahed, Sepideh; Jahanban-Esfahlan, Ali; Jaymand, Mehdi; Majdi, Hasan; Amoozgar, Zohreh; Chitkushev, L. T. et al. (2020). "Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression". Frontiers in Cell and Developmental Biology 8: 229. doi:10.3389/fcell.2020.00229. ISSN 2296-634X. PMID 32509768.
- ↑ Dubois, Alain (2007-08-01). "Naming taxa from cladograms: some confusions, misleading statements, and necessary clarifications" (in en). Cladistics 23 (4): 390–402. doi:10.1111/j.1096-0031.2007.00151.x. ISSN 0748-3007. PMID 34905840. https://onlinelibrary.wiley.com/doi/10.1111/j.1096-0031.2007.00151.x.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ d'Huy 2012a, b; d'Huy 2013a, b, c, d
- ↑ Ross and al. 2013
- ↑ Tehrani 2013
- ↑ "Canterbury Tales Project". http://www.canterburytalesproject.org.
- ↑ 52.0 52.1 Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
- ↑ Lua error: Internal error: The interpreter exited with status 1.
Lua error: Internal error: The interpreter exited with status 1.
Bibliography
- Adrain, Jonathan M.; Edgecombe, Gregory D.; Lieberman, Bruce S. (2002), Fossils, Phylogeny, and Form: An Analytical Approach, New York: Kluwer Academic, ISBN 978-0-306-46721-9, https://books.google.com/books?id=fpnV7NTcnuAC&pg=PA56, retrieved 2012-08-15
- Baron, C.; Høeg, J.T. (2005), "Gould, Scharm and the paleontologocal perspective in evolutionary biology", in Koenemann, S.; Jenner, R.A., Crustacea and Arthropod Relationships, CRC Press, pp. 3–14, ISBN 978-0-8493-3498-6, https://books.google.com/books?id=LalmQ4346O0C&q=Nielsen,+C.+2001+%22Animal+evolution%22+Chelicerata, retrieved 2008-10-15
- Benton, M. J. (2000), "Stems, nodes, crown clades, and rank-free lists: is Linnaeus dead?", Biological Reviews 75 (4): 633–648, doi:10.1111/j.1469-185X.2000.tb00055.x, PMID 11117201, http://palaeo.gly.bris.ac.uk/Benton/reprints/2000phylocode.pdf, retrieved 2 October 2011
- Benton, M. J. (2004), Vertebrate Palaeontology (3rd ed.), Oxford: Blackwell Science, ISBN 978-0-632-05637-8
- Brinkman, Fiona S.L.; Leipe, Detlef D. (2001), Baxevanis, Andreas D.; Ouellette, B.F. Francis, eds., Phylogenetic analysis (2nd ed.), pp. 323–358, http://www.bioon.com/book/biology/Bioinformatics/chapter-14.pdf, retrieved 2013-10-19
- Cain, A. J.; Harrison, G. A. (1960), "Phyletic weighting", Proceedings of the Zoological Society of London 35: 1–31, doi:10.1111/j.1469-7998.1960.tb05828.x
- Cuénot, Lucien (1940), "Remarques sur un essai d'arbre généalogique du règne animal", Comptes Rendus de l'Académie des Sciences de Paris 210: 23–27. Available free online at Gallica (No direct URL). This is the paper credited by Lua error: Internal error: The interpreter exited with status 1. for the first use of the term 'clade'.
- Dupuis, Claude (1984), "Willi Hennig's impact on taxonomic thought", Annual Review of Ecology and Systematics 15: 1–24, doi:10.1146/annurev.ecolsys.15.1.1, ISSN 0066-4162.
- Farris, James S. (1977), "On the phenetic approach to vertebrate classification", in Hecht, M. K.; Goody, P. C.; Hecht, B. M., Major Patterns in Vertebrate Evolution, Plenum, New York, pp. 823–850
- Farris, James S. (1979a), "On the naturalness of phylogenetic classification", Systematic Zoology 28 (2): 200–214, doi:10.2307/2412523
- Farris, James S. (1979b), "The information content of the phylogenetic system", Systematic Zoology 28 (4): 483–519, doi:10.2307/2412562
- Farris, James S. (1980), "The efficient diagnoses of the phylogenetic system", Systematic Zoology 29 (4): 386–401, doi:10.2307/2992344
- Farris, James S. (1983), "The logical basis of phylogenetic analysis", in Platnick, Norman I.; Funk, Vicki A., Advances in Cladistics, vol. 2, Columbia University Press, New York, pp. 7–36
- Fraix-Burnet, D.; Choler, P.; Douzery, E.J.P.; Verhamme, A. (2006), "Astrocladistics: A Phylogenetic Analysis of Galaxy Evolution II. Formation and Diversification of Galaxies", Journal of Classification 23 (1): 57–78, doi:10.1007/s00357-006-0004-4, Bibcode: 2006JClas..23...57F
- Hennig, Willi (1966), Phylogenetic systematics (tr. D. Dwight Davis and Rainer Zangerl), Urbana, IL: Univ. of Illinois Press (reprinted 1979 and 1999), ISBN 978-0-252-06814-0
- Hennig, Willi (1975), "'Cladistic analysis or cladistic classification?': a reply to Ernst Mayr", Systematic Zoology 24 (2): 244–256, doi:10.2307/2412765, http://joelvelasco.net/teaching/systematics/Hennig%201974%20-%20Cladistic%20Analysis%20or%20Cladistic%20Classification.pdf, responding to Lua error: Internal error: The interpreter exited with status 1..
- Hennig, Willi (1999), Phylogenetic systematics (3rd edition of 1966 book), Urbana: University of Illinois Press, ISBN 978-0-252-06814-0 Translated from manuscript in German eventually published in 1982 (Phylogenetische Systematik, Verlag Paul Parey, Berlin).
- Hull, David (1988), Science as a Process, University of Chicago Press, ISBN 978-0-226-36051-5
- James, Frances C.; Pourtless IV, John A. (2009), Cladistics and the Origin of Birds: A Review and Two New Analyses, Ornithological Monographs, No. 66, American Ornithologists' Union, ISBN 978-0-943610-85-6, http://www.bio.fsu.edu/James/Ornithological%20Monographs%202009.pdf, retrieved 2010-12-14
- d'Huy, Julien (2012a). "Un ours dans les étoiles: recherche phylogénétique sur un mythe préhistorique". Préhistoire du Sud-Ouest 20 (1): 91–106. https://www.academia.edu/3226058.
- d'Huy, Julien (2012b), "Le motif de Pygmalion : origine afrasienne et diffusion en Afrique". Sahara, 23: 49-59 [1].
- d'Huy, Julien (2013a), "Polyphemus (Aa. Th. 1137)." "A phylogenetic reconstruction of a prehistoric tale". Nouvelle Mythologie Comparée / New Comparative Mythology 1, [2]
- d'Huy, Julien (2013b). "A phylogenetic approach of mythology and its archaeological consequences". Rock Art Research 30 (1): 115–118. https://www.academia.edu/3366931.
- d'Huy, Julien (2013c) "Les mythes évolueraient par ponctuations". Mythologie française, 252, 2013c: 8-12. [3]
- d'Huy, Julien (2013d) "A Cosmic Hunt in the Berber sky : a phylogenetic reconstruction of Palaeolithic mythology". Les Cahiers de l'AARS, 15, 2013d: 93-106. [4]
- Jerison, Harry J. (2003), "On Theory in Comparative Psychology", in Sternberg, Robert J.; Kaufman, James C., The Evolution of Intelligence, Mahwah, NJ: Lawrence Erlbaum Associates, Inc., ISBN 978-0-12-385250-2
- Lipo, Carl; O'Brien, Michael J.; Collard, Mark et al., eds. (2006), Mapping Our Ancestors: Phylogenetic Approaches in Anthropology and Prehistory, Piscataway: Transaction Publishers, ISBN 978-0-202-30751-0
- Maas, Philipp (2010–2011), Jürgen, Hanneder; Maas, Philipp, eds., "Computer Aided Stemmatics – The Case of Fifty-Two Text Versions of Carakasasaṃhitā Vimānasthāna 8.67-157", Wiener Zeitschrift für die Kunde Südasiens 52–53: 63–120, doi:10.1553/wzks2009-2010s63
- Mace, Ruth; Clare, Clare J.; Shennan, Stephen, eds. (2005), The Evolution of Cultural Diversity: A Phylogenetic Approach, Portland: Cavendish Press, ISBN 978-1-84472-099-6
- Mayr, Ernst (1974), "Cladistic analysis or cladistic classification?", Zeitschrift für Zoologische Systematik und Evolutionsforschung 12: 94–128, doi:10.1111/j.1439-0469.1974.tb00160.x, http://courses.cit.cornell.edu/jdv55/teaching/systematics/mayr%2074%20-%20cladistic%20analysis%20or%20cladistic%20classification.pdf
- Mayr, Ernst (1976), Evolution and the diversity of life (Selected essays), Cambridge, Massachusetts: Harvard University Press, ISBN 978-0-674-27105-0 Reissued 1997 in paperback. Includes a reprint of Mayr's 1974 anti-cladistics paper at pp. 433–476, "Cladistic analysis or cladistic classification." This is the paper to which Lua error: Internal error: The interpreter exited with status 1. is a response.
- Mayr, Ernst (1978), "Origin and history of some terms in systematic and evolutionary biology", Systematic Zoology 27 (1): 83–88, doi:10.2307/2412818.
- Mayr, Ernst (1982), The growth of biological thought: diversity, evolution and inheritance, Cambridge, Massachusetts: Harvard University Press, ISBN 978-0-674-36446-2
- Oppenheimer, Stephen (2006), The Origins of the British, London: Robinson, ISBN 978-0-7867-1890-0, https://archive.org/details/isbn_9780786718900
- Patterson, Colin (1982), "Morphological characters and homology", in Joysey, Kenneth A; Friday, A. E., Problems in Phylogenetic Reconstruction, Systematics Association Special Volume 21, London: Academic Press, ISBN 978-0-12-391250-3.
- Patterson, Colin (1988), "Homology in classical and molecular biology", Molecular Biology and Evolution 5 (6): 603–625, doi:10.1093/oxfordjournals.molbev.a040523, PMID 3065587
- de Pinna, M.G.G (1991), "Concepts and tests of homology in the cladistic paradigm", Cladistics 7 (4): 367–394, doi:10.1111/j.1096-0031.1991.tb00045.x, http://www.ib.usp.br/hennig/depinna1991.pdf, retrieved 24 October 2017
- de Queiroz, K.; Gauthier, J. (1992), "Phylogenetic taxonomy", Annual Review of Ecology and Systematics 23: 449–480, doi:10.1146/annurev.ecolsys.23.1.449, http://www.faculty.biol.ttu.edu/strauss/Phylogenetics/Readings/deQueirozGauthier1992.pdf, retrieved 28 July 2012
- Robinson, Peter M.W.; O’Hara, Robert J. (1996), "Cladistic analysis of an Old Norse manuscript tradition", Research in Humanities Computing 4: 115–137, http://rjohara.net/cv/1996-rhc, retrieved 2010-12-13
- Ross, Robert M.; Greenhill, Simon J.; Atkinson, Quentin D. (2013). "Population structure and cultural geography of a folktale in Europe". Proceedings of the Royal Society B: Biological Sciences 280 (1756): 20123065. doi:10.1098/rspb.2012.3065. PMID 23390109.
- Schuh, Randall T.; Brower, Andrew V.Z. (2009), Biological Systematics: Principles and Applications (2nd ed.), Cornell University Press, ISBN 978-0-8014-4799-0
- Taylor, Mike (2003), What do terms like monophyletic, paraphyletic and polyphyletic mean?, http://www.miketaylor.org.uk/dino/faq/s-class/phyletic/, retrieved 2010-12-13
- Tehrani, Jamshid J., 2013, "The Phylogeny of Little Red Riding Hood", PLOS ONE, 13 November.[5]
- Tremblay, Frederic (2013), "Nicolai Hartmann and the Metaphysical Foundation of Phylogenetic Systematics", Biological Theory 7 (1): 56–68, doi:10.1007/s13752-012-0077-8, https://philarchive.org/rec/TRENHA
- Weygoldt, P. (February 1998), "Evolution and systematics of the Chelicerata", Experimental and Applied Acarology 22 (2): 63–79, doi:10.1023/A:1006037525704
- Wheeler, Quentin (2000), Species Concepts and Phylogenetic Theory: A Debate, Columbia University Press, ISBN 978-0-231-10143-1
- Wiley, E.O.; Siegel-Causey, D.; Brooks, D.R.; Funk, V.A. (1991), "Chapter 1 Introduction, terms and concepts", The Compleat Cladist: A Primer of Phylogenetic Procedures, The University of Kansas Museum of Natural History, ISBN 978-0-89338-035-9, http://www.amnh.org/learn/pd/fish_2/pdf/compleat_cladist.pdf, retrieved 2010-12-13
- Williams, P.A. (1992), "Confusion in cladism", Synthese 01 (1–2): 135–152, doi:10.1007/BF00484973
External links
- OneZoom: Tree of Life – all living species as intuitive and zoomable fractal explorer (responsive design)
- Willi Hennig Society
- Cladistics (scholarly journal of the Willi Hennig Society)
- Collins, Allen G.; Guralnick, Rob; Smith, Dave (1994–2005). "Journey into Phylogenetic Systematics". University of California Museum of Paleontology. http://www.ucmp.berkeley.edu/clad/clad4.html.
- Felsenstein, Joe. "Phylogeny Programs". Seattle: University of Washington. http://evolution.gs.washington.edu/phylip/software.html.
- O'Neil, Dennis (1998–2008). "Classification of Living Things". San Marcos CA: Palomar College. http://anthro.palomar.edu/animal/default.htm.
- Robinson, Peter; O'Hara, Robert J. (1992). "Report on the Textual Criticism Challenge 1991". rjohara.net. http://rjohara.net/darwin/files/bmcr.
- Theobald, Douglas (1999–2004). "Phylogenetics Primer". The TalkOrigins Archive. http://www.talkorigins.org/faqs/comdesc/phylo.html.
Lua error: Internal error: The interpreter exited with status 1.
Lua error: Internal error: The interpreter exited with status 1.