Biology:DfrA-dnaX RNA motif
| dfrA-dnaX | |
|---|---|
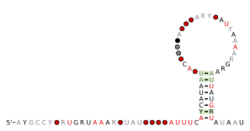 Consensus secondary structure and sequence conservation of dfrA-dnaX RNA | |
| Identifiers | |
| Symbol | dfrA-dnaX |
| Rfam | RF02952 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The dfrA-dnaX RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] dfrA-dnaX motifs are found in Parvimonas.
It is ambiguous whether dfrA-dnaX RNAs function as cis-regulatory elements or not. dfrA-dnaX RNAs are apparently located in the 5' untranslated regions (UTRs) of a variety of non-homologous genes. They are almost never (<1%) in locations that are unlikely to correspond to a 5' UTR. These data suggest a role as a cis regulator. However, the motif consists entirely of a hairpin whose terminal loop is quite large, and the motif is not well conserved. This lack of conservation is surprising in view of the narrow range of organisms with the motif. Therefore, it was proposed that it is possible that the motif serves another more general role, such as Rho-independent transcription termination.[1]
References
- ↑ 1.0 1.1 "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |

