Biology:Förster resonance energy transfer
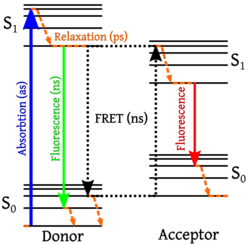
Förster resonance energy transfer (FRET), fluorescence resonance energy transfer, resonance energy transfer (RET) or electronic energy transfer (EET) is a mechanism describing energy transfer between two light-sensitive molecules (chromophores).[1] A donor chromophore, initially in its electronic excited state, may transfer energy to an acceptor chromophore through nonradiative dipole–dipole coupling.[2] The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET extremely sensitive to small changes in distance.[3][4]
Measurements of FRET efficiency can be used to determine if two fluorophores are within a certain distance of each other.[5] Such measurements are used as a research tool in fields including biology and chemistry.
FRET is analogous to near-field communication, in that the radius of interaction is much smaller than the wavelength of light emitted. In the near-field region, the excited chromophore emits a virtual photon that is instantly absorbed by a receiving chromophore. These virtual photons are undetectable, since their existence violates the conservation of energy and momentum, and hence FRET is known as a radiationless mechanism. Quantum electrodynamical calculations have been used to determine that radiationless (FRET) and radiative energy transfer are the short- and long-range asymptotes of a single unified mechanism.[6][7][8]
Terminology

Förster resonance energy transfer is named after the German scientist Theodor Förster.[9] When both chromophores are fluorescent, the term "fluorescence resonance energy transfer" is often used instead, although the energy is not actually transferred by fluorescence.[10][11] In order to avoid an erroneous interpretation of the phenomenon that is always a nonradiative transfer of energy (even when occurring between two fluorescent chromophores), the name "Förster resonance energy transfer" is preferred to "fluorescence resonance energy transfer"; however, the latter enjoys common usage in scientific literature.[12] FRET is not restricted to fluorescence and occurs in connection with phosphorescence as well.[10]
Theoretical basis
The FRET efficiency () is the quantum yield of the energy-transfer transition, i.e. the probability of energy-transfer event occurring per donor excitation event:[13]
where the radiative decay rate of the donor, is the rate of energy transfer, and the rates of any other de-excitation pathways excluding energy transfers to other acceptors.[14][15]
The FRET efficiency depends on many physical parameters[16] that can be grouped as: 1) the distance between the donor and the acceptor (typically in the range of 1–10 nm), 2) the spectral overlap of the donor emission spectrum and the acceptor absorption spectrum, and 3) the relative orientation of the donor emission dipole moment and the acceptor absorption dipole moment.
depends on the donor-to-acceptor separation distance with an inverse 6th-power law due to the dipole–dipole coupling mechanism:
with being the Förster distance of this pair of donor and acceptor, i.e. the distance at which the energy transfer efficiency is 50%.[14] The Förster distance depends on the overlap integral of the donor emission spectrum with the acceptor absorption spectrum and their mutual molecular orientation as expressed by the following equation all in SI units:[17][18][19]
where is the fluorescence quantum yield of the donor in the absence of the acceptor, is the dipole orientation factor, is the refractive index of the medium, is the Avogadro constant, and is the spectral overlap integral calculated as
where is the donor emission spectrum, is the donor emission spectrum normalized to an area of 1, and is the acceptor molar extinction coefficient, normally obtained from an absorption spectrum.[20] The orientation factor κ is given by
where denotes the normalized transition dipole moment of the respective fluorophore, and denotes the normalized inter-fluorophore displacement.[21] = 2/3 is often assumed. This value is obtained when both dyes are freely rotating and can be considered to be isotropically oriented during the excited-state lifetime. If either dye is fixed or not free to rotate, then = 2/3 will not be a valid assumption. In most cases, however, even modest reorientation of the dyes results in enough orientational averaging that = 2/3 does not result in a large error in the estimated energy-transfer distance due to the sixth-power dependence of on . Even when is quite different from 2/3, the error can be associated with a shift in , and thus determinations of changes in relative distance for a particular system are still valid. Fluorescent proteins do not reorient on a timescale that is faster than their fluorescence lifetime. In this case 0 ≤ ≤ 4.[20]
The units of the data are usually not in SI units. Using the original units to calculate the Förster distance is often more convenient. For example, the wavelength is often in unit nm and the extinction coefficient is often in unit , where is concentration . obtained from these units will have unit . To use unit Å () for the , the equation is adjusted to[17][22][23][24]
- (Å)
For time-dependent analyses of FRET, the rate of energy transfer () can be used directly instead:[17]
where is the donor's fluorescence lifetime in the absence of the acceptor.
The FRET efficiency relates to the quantum yield and the fluorescence lifetime of the donor molecule as follows:[25]
where and are the donor fluorescence lifetimes in the presence and absence of an acceptor respectively, or as
where and are the donor fluorescence intensities with and without an acceptor respectively.
Experimental confirmation of the FRET theory
The inverse sixth-power distance dependence of Förster resonance energy transfer was experimentally confirmed by Wilchek, Edelhoch and Brand[26] using tryptophyl peptides. Stryer, Haugland and Yguerabide[27][citation needed][28] also experimentally demonstrated the theoretical dependence of Förster resonance energy transfer on the overlap integral by using a fused indolosteroid as a donor and a ketone as an acceptor. Calculations on FRET distances of some example dye-pairs can be found here.[22][24] However, a lot of contradictions of special experiments with the theory was observed under complicated environment when the orientations and quantum yields of the molecules are difficult to estimate.[29]
Methods to measure FRET efficiency
In fluorescence microscopy, fluorescence confocal laser scanning microscopy, as well as in molecular biology, FRET is a useful tool to quantify molecular dynamics in biophysics and biochemistry, such as protein-protein interactions, protein–DNA interactions, and protein conformational changes. For monitoring the complex formation between two molecules, one of them is labeled with a donor and the other with an acceptor. The FRET efficiency is measured and used to identify interactions between the labeled complexes. There are several ways of measuring the FRET efficiency by monitoring changes in the fluorescence emitted by the donor or the acceptor.[30]
Sensitized emission
One method of measuring FRET efficiency is to measure the variation in acceptor emission intensity.[18] When the donor and acceptor are in proximity (1–10 nm) due to the interaction of the two molecules, the acceptor emission will increase because of the intermolecular FRET from the donor to the acceptor. For monitoring protein conformational changes, the target protein is labeled with a donor and an acceptor at two loci. When a twist or bend of the protein brings the change in the distance or relative orientation of the donor and acceptor, FRET change is observed. If a molecular interaction or a protein conformational change is dependent on ligand binding, this FRET technique is applicable to fluorescent indicators for the ligand detection.
Photobleaching FRET
FRET efficiencies can also be inferred from the photobleaching rates of the donor in the presence and absence of an acceptor.[18] This method can be performed on most fluorescence microscopes; one simply shines the excitation light (of a frequency that will excite the donor but not the acceptor significantly) on specimens with and without the acceptor fluorophore and monitors the donor fluorescence (typically separated from acceptor fluorescence using a bandpass filter) over time. The timescale is that of photobleaching, which is seconds to minutes, with fluorescence in each curve being given by
where is the photobleaching decay time constant and depends on whether the acceptor is present or not. Since photobleaching consists in the permanent inactivation of excited fluorophores, resonance energy transfer from an excited donor to an acceptor fluorophore prevents the photobleaching of that donor fluorophore, and thus high FRET efficiency leads to a longer photobleaching decay time constant:
where and are the photobleaching decay time constants of the donor in the presence and in the absence of the acceptor respectively. (Notice that the fraction is the reciprocal of that used for lifetime measurements).
This technique was introduced by Jovin in 1989.[31] Its use of an entire curve of points to extract the time constants can give it accuracy advantages over the other methods. Also, the fact that time measurements are over seconds rather than nanoseconds makes it easier than fluorescence lifetime measurements, and because photobleaching decay rates do not generally depend on donor concentration (unless acceptor saturation is an issue), the careful control of concentrations needed for intensity measurements is not needed. It is, however, important to keep the illumination the same for the with- and without-acceptor measurements, as photobleaching increases markedly with more intense incident light.
Lifetime measurements
FRET efficiency can also be determined from the change in the fluorescence lifetime of the donor.[18] The lifetime of the donor will decrease in the presence of the acceptor. Lifetime measurements of the FRET-donor are used in fluorescence-lifetime imaging microscopy (FLIM).
Single-molecule FRET (smFRET)
smFRET is a group of methods using various microscopic techniques to measure a pair of donor and acceptor fluorophores that are excited and detected at the single molecule level. In contrast to "ensemble FRET" or "bulk FRET" which provides the FRET signal of a high number of molecules, single-molecule FRET is able to resolve the FRET signal of each individual molecule. The variation of the smFRET signal is useful to reveal kinetic information that an ensemble measurement cannot provide, especially when the system is under equilibrium. Heterogeneity among different molecules can also be observed. This method has been applied in many measurements of biomolecular dynamics such as DNA/RNA/protein folding/unfolding and other conformational changes, and intermolecular dynamics such as reaction, binding, adsorption, and desorption that are particularly useful in chemical sensing, bioassays, and biosensing.
Fluorophores used for FRET
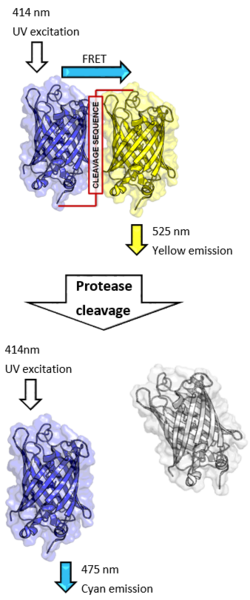
CFP-YFP pairs
One common pair fluorophores for biological use is a cyan fluorescent protein (CFP) – yellow fluorescent protein (YFP) pair.[32] Both are color variants of green fluorescent protein (GFP). Labeling with organic fluorescent dyes requires purification, chemical modification, and intracellular injection of a host protein. GFP variants can be attached to a host protein by genetic engineering which can be more convenient. Additionally, a fusion of CFP and YFP ("tandem-dimer") linked by a protease cleavage sequence can be used as a cleavage assay.[33]
BRET
A limitation of FRET performed with fluorophore donors is the requirement for external illumination to initiate the fluorescence transfer, which can lead to background noise in the results from direct excitation of the acceptor or to photobleaching. To avoid this drawback, bioluminescence resonance energy transfer (or BRET) has been developed.[34][35] This technique uses a bioluminescent luciferase (typically the luciferase from Renilla reniformis) rather than CFP to produce an initial photon emission compatible with YFP.
BRET has also been implemented using a different luciferase enzyme, engineered from the deep-sea shrimp Oplophorus gracilirostris. This luciferase is smaller (19 kD) and brighter than the more commonly used luciferase from Renilla reniformis,[36][37][38][39] and has been named NanoLuc[40] or NanoKAZ.[41] Promega has developed a patented substrate for NanoLuc called furimazine,[42][40] though other valuables coelenterazine substrates for NanoLuc have also been published.[41][43] A split-protein version of NanoLuc developed by Promega[44] has also been used as a BRET donor in experiments measuring protein-protein interactions.[45]
Homo-FRET
In general, "FRET" refers to situations where the donor and acceptor proteins (or "fluorophores") are of two different types. In many biological situations, however, researchers might need to examine the interactions between two, or more, proteins of the same type—or indeed the same protein with itself, for example if the protein folds or forms part of a polymer chain of proteins[46] or for other questions of quantification in biological cells[47] or in vitro experiments.[48]
Obviously, spectral differences will not be the tool used to detect and measure FRET, as both the acceptor and donor protein emit light with the same wavelengths. Yet researchers can detect differences in the polarisation between the light which excites the fluorophores and the light which is emitted, in a technique called FRET anisotropy imaging; the level of quantified anisotropy (difference in polarisation between the excitation and emission beams) then becomes an indicative guide to how many FRET events have happened.[49]
In the field of nano-photonics, FRET can be detrimental if it funnels excitonic energy to defect sites, but it is also essential to charge collection in organic and quantum-dot-sensitized solar cells, and various FRET-enabled strategies have been proposed for different opto-electronic devices. It is then essential to understand how isolated nano-emitters behave when they are stacked in a dense layer. Nanoplatelets are especially promising candidates for strong homo-FRET exciton diffusion because of their strong in-plane dipole coupling and low Stokes shift.[50] Fluorescence microscopy study of such single chains demonstrated that energy transfer by FRET between neighbor platelets causes energy to diffuse over a typical 500-nm length (about 80 nano emitters), and the transfer time between platelets is on the order of 1 ps.[51]
Others
Various compounds beside fluorescent proteins.[52]
Applications
The applications of fluorescence resonance energy transfer (FRET) have expanded tremendously in the last 25 years, and the technique has become a staple in many biological and biophysical fields. FRET can be used as a spectroscopic ruler to measure distance and detect molecular interactions in a number of systems and has applications in biology and biochemistry.[28][53]
Proteins
FRET is often used to detect and track interactions between proteins.[54][55][56][57] Additionally, FRET can be used to measure distances between domains in a single protein by tagging different regions of the protein with fluorophores and measuring emission to determine distance. This provides information about protein conformation, including secondary structures and protein folding.[58][59] This extends to tracking functional changes in protein structure, such as conformational changes associated with myosin activity.[60] Applied in vivo, FRET has been used to detect the location and interactions of cellular structures including integrins and membrane proteins.[61]
Membranes
FRET can be used to observe membrane fluidity, movement and dispersal of membrane proteins, membrane lipid-protein and protein-protein interactions, and successful mixing of different membranes.[62] FRET is also used to study formation and properties of membrane domains and lipid rafts in cell membranes[63] and to determine surface density in membranes.[64]
Chemosensor

FRET-based probes can detect the presence of various molecules: the probe's structure is affected by small molecule binding or activity, which can turn the FRET system on or off. This is often used to detect anions, cations, small uncharged molecules, and some larger biomacromolecules as well. Similarly, FRET systems have been designed to detect changes in the cellular environment due to such factors as pH, hypoxia, or mitochondrial membrane potential.[65]
Signaling pathways
Another use for FRET is in the study of metabolic or signaling pathways.[66] For example, FRET and BRET have been used in various experiments to characterize G-protein coupled receptor activation and consequent signaling mechanisms.[67] Other examples include the use of FRET to analyze such diverse processes as bacterial chemotaxis[68] and caspase activity in apoptosis.[69]
Proteins and nucleotides folding kinetics
Proteins, DNAs, RNAs, and other polymer folding dynamics have been measured using FRET. Usually, these systems are under equilibrium whose kinetics is hidden. However, they can be measured by measuring single-molecule FRET with proper placement of the acceptor and donor dyes on the molecules. See single-molecule FRET for a more detailed description.
Other applications
In addition to common uses previously mentioned, FRET and BRET are also effective in the study of biochemical reaction kinetics.[70] FRET is increasingly used for monitoring pH dependent assembly and disassembly and is valuable in the analysis of nucleic acids encapsulation.[71][72][73][74] This technique can be used to determine factors affecting various types of nanoparticle formation[75][76] as well as the mechanisms and effects of nanomedicines.[77]
Other methods
A different, but related, mechanism is Dexter electron transfer.
An alternative method to detecting protein–protein proximity is the bimolecular fluorescence complementation (BiFC), where two parts of a fluorescent protein are each fused to other proteins. When these two parts meet, they form a fluorophore on a timescale of minutes or hours.[78]
See also
- Dexter electron transfer
- Förster coupling
- Surface energy transfer
- Time-resolved fluorescence energy transfer
References
- ↑ Cheng, Ping-Chin (2006). "The Contrast Formation in Optical Microscopy". in Pawley, James B.. Handbook of Biological Confocal Microscopy (3rd ed.). New York, NY: Springer. pp. 162–206. doi:10.1007/978-0-387-45524-2_8. ISBN 978-0-387-25921-5. https://books.google.com/books?id=E2maxdEXFNoC&pg=PA162.
- ↑ Helms, Volkhard (2008). "Fluorescence Resonance Energy Transfer". Principles of Computational Cell Biology. Weinheim: Wiley-VCH. p. 202. ISBN 978-3-527-31555-0. https://books.google.com/books?id=-Tavvybv5UwC&pg=PA202.
- ↑ Harris, Daniel C. (2010). "Applications of Spectrophotometry". Quantitative Chemical Analysis (8th ed.). New York: W. H. Freeman and Co.. pp. 419–44. ISBN 978-1-4292-1815-3. https://books.google.com/books?id=kIgLJ1De_jwC&pg=PA419.
- ↑ Schneckenburger, Herbert (2019-11-27). "Förster resonance energy transfer–what can we learn and how can we use it?". Methods and Applications in Fluorescence 8 (1): 013001. doi:10.1088/2050-6120/ab56e1. ISSN 2050-6120. PMID 31715588.
- ↑ "Spectroscopy-based quantitative fluorescence resonance energy transfer analysis". Ion Channels. Methods in Molecular Biology. 337. Humana Press. 2006. pp. 65–77. doi:10.1385/1-59745-095-2:65. ISBN 978-1-59745-095-9. https://books.google.com/books?id=Q2k-T_1DPcwC&pg=PA65.
- ↑ Andrews, David L. (1989). "A unified theory of radiative and radiationless molecular energy transfer". Chemical Physics 135 (2): 195–201. doi:10.1016/0301-0104(89)87019-3. Bibcode: 1989CP....135..195A. https://ueaeprints.uea.ac.uk/56540/1/050.pdf.
- ↑ Andrews, David L; Bradshaw, David S (2004). "Virtual photons, dipole fields and energy transfer: A quantum electrodynamical approach". European Journal of Physics 25 (6): 845–858. doi:10.1088/0143-0807/25/6/017. https://ueaeprints.uea.ac.uk/10692/4/0143_0807_25_6_017.pdf.
- ↑ Jones, Garth A; Bradshaw, David S (2019). "Resonance energy transfer: From fundamental theory to recent applications". Frontiers in Physics 7: 100. doi:10.3389/fphy.2019.00100. Bibcode: 2019FrP.....7..100J.
- ↑ Förster, Theodor (1948). "Zwischenmolekulare Energiewanderung und Fluoreszenz" (in de). Annalen der Physik 437 (1–2): 55–75. doi:10.1002/andp.19484370105. Bibcode: 1948AnP...437...55F.
- ↑ 10.0 10.1 Valeur, Bernard; Berberan-Santos, Mario (2012). "Excitation Energy Transfer". Molecular Fluorescence: Principles and Applications, 2nd ed.. Weinheim: Wiley-VCH. pp. 213–261. doi:10.1002/9783527650002.ch8. ISBN 978-3-527-32837-6.
- ↑ FRET microscopy tutorial from Olympus
- ↑ Glossary of Terms Used in Photochemistry (3rd ed.). IUPAC. 2007. p. 340.
- ↑ Moens, Pierre. "Fluorescence Resonance Energy Transfer spectroscopy". http://www.anatomy.usyd.edu.au/mru/fret/abot.html#frete.
- ↑ 14.0 14.1 Schaufele, Fred; Demarco, Ignacio; Day, Richard N. (2005). "FRET Imaging in the Wide-Field Microscope". in Periasamy, Ammasi; Day, Richard. Molecular Imaging: FRET Microscopy and Spectroscopy. Oxford: Oxford University Press. pp. 72–94. doi:10.1016/B978-019517720-6.50013-4. ISBN 978-0-19-517720-6. https://books.google.com/books?id=K0aawJ6sX-sC&pg=PA72.
- ↑ "Single-molecule three-color FRET with both negligible spectral overlap and long observation time". PLOS ONE 5 (8): e12270. August 2010. doi:10.1371/journal.pone.0012270. PMID 20808851. Bibcode: 2010PLoSO...512270L.
- ↑ C. King; B. Barbiellini; D. Moser; V. Renugopalakrishnan (2012). "Exactly soluble model of resonant energy transfer between molecules". Physical Review B 85 (12): 125106. doi:10.1103/PhysRevB.85.125106. Bibcode: 2012PhRvB..85l5106K.
- ↑ 17.0 17.1 17.2 Förster, Th. (1965). "Delocalized Excitation and Excitation Transfer". in Sinanoglu, Oktay. Modern Quantum Chemistry. Istanbul Lectures. Part III: Action of Light and Organic Crystals. 3. New York and London: Academic Press. pp. 93–137. http://www.quantum-chemistry-history.com/Sina_Dat/BOOKIstaLec/IstaLec1.htm. Retrieved 2011-06-22.
- ↑ 18.0 18.1 18.2 18.3 Clegg, Robert (2009). "Förster resonance energy transfer—FRET: what is it, why do it, and how it's done". in Gadella, Theodorus W. J.. FRET and FLIM Techniques. Laboratory Techniques in Biochemistry and Molecular Biology. 33. Elsevier. pp. 1–57. doi:10.1016/S0075-7535(08)00001-6. ISBN 978-0-08-054958-3. https://books.google.com/books?id=uHvqu4hLhH8C&pg=PA1.
- ↑ http://spie.org/samples/PM194.pdf [bare URL PDF]
- ↑ 20.0 20.1 Demchenko, Alexander P. (2008). "Fluorescence Detection Techniques". Introduction to Fluorescence Sensing. Dordrecht: Springer. pp. 65–118. doi:10.1007/978-1-4020-9003-5_3. ISBN 978-1-4020-9002-8. https://books.google.com/books?id=wMARxPxkE7EC&pg=PA65.
- ↑ VanDerMeer, B. Wieb (2020). "Kappaphobia is the elephant in the fret room" (in en). Methods and Applications in Fluorescence 8 (3): 030401. doi:10.1088/2050-6120/ab8f87. ISSN 2050-6120. PMID 32362590. Bibcode: 2020MApFl...8c0401V.
- ↑ 22.0 22.1 "FPbase FRET Calculator". https://www.fpbase.org/fret/.
- ↑ "Using patterned arrays of metal nanoparticles to probe plasmon enhanced luminescence of CdSe quantum dots (supporting information)" (in en). ACS Nano 3 (7): 1735–1744. 2009. doi:10.1021/nn900317n. PMID 19499906. https://pubs.acs.org/doi/10.1021/nn900317n.
- ↑ 24.0 24.1 "Resonance Energy Transfer: Methods and Applications" (in en). Analytical Biochemistry 218 (1): 1–13. 1994. doi:10.1006/abio.1994.1134. PMID 8053542.
- ↑ Majoul, Irina; Jia, Yiwei; Duden, Rainer (2006). "Practical Fluorescence Resonance Energy Transfer or Molecular Nanobioscopy of Living Cells". in Pawley, James B.. Handbook of Biological Confocal Microscopy (3rd ed.). New York, NY: Springer. pp. 788–808. doi:10.1007/978-0-387-45524-2_45. ISBN 978-0-387-25921-5. https://archive.org/details/handbookbiologic00pawl.
- ↑ "Fluorescence studies with tryptophyl peptides". Biochemistry 6 (2): 547–59. February 1967. doi:10.1021/bi00854a024. PMID 6047638.
- ↑ Lakowicz, Joseph R., ed (1991). Principles. New York: Plenum Press. p. 172. ISBN 978-0-306-43875-2.
- ↑ 28.0 28.1 Lakowicz, Joseph R. (1999). Principles of fluorescence spectroscopy (2nd ed.). New York, NY: Kluwer Acad./Plenum Publ.. pp. 374–443. ISBN 978-0-306-46093-7. https://archive.org/details/principlesfluore00lako.
- ↑ "Energy Transfer in Macromolecules, SPIE". Photonics of Biopolymers. Springer. 1997.
- ↑ "Fluorescence Resonance Energy Transfer Protocol". Wellcome Trust. http://coil.bio.ed.ac.uk/Protocols/FRET.htm.
- ↑ Szöllősi, János; Alexander, Denis R. (2007). "The Application of Fluorescence Resonance Energy Transfer to the Investigation of Phosphatases". in Klumpp, Susanne; Krieglstein, Josef. Protein Phosphatases. Methods in Enzymology. 366. Amsterdam: Elsevier. pp. 203–24. doi:10.1016/S0076-6879(03)66017-9. ISBN 978-0-12-182269-9.
- ↑ "Fluorescence resonance energy transfer microscopy: a mini review". Journal of Biomedical Optics 6 (3): 287–91. July 2001. doi:10.1117/1.1383063. PMID 11516318. Bibcode: 2001JBO.....6..287P. https://pdfs.semanticscholar.org/3b4d/4d92c44059eabee448451f83bc7ac71f9630.pdf.
- ↑ "Evolutionary optimization of fluorescent proteins for intracellular FRET". Nature Biotechnology 23 (3): 355–60. March 2005. doi:10.1038/nbt1066. PMID 15696158.
- ↑ Bevan, Nicola; Rees, Stephen (2006). "Pharmaceutical Applications of GFP and RCFP". in Chalfie, Martin; Kain, Steven R.. Green Fluorescent Protein: Properties, Applications and Protocols. Methods of Biochemical Analysis. 47 (2nd ed.). Hoboken, NJ: John Wiley & Sons. pp. 361–90. doi:10.1002/0471739499.ch16. ISBN 978-0-471-73682-0. https://books.google.com/books?id=v8Y4zrEofpIC&pg=PA361.
- ↑ "Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET)". Nature Methods 3 (3): 165–74. March 2006. doi:10.1038/nmeth841. PMID 16489332.
- ↑ "Enabling systematic interrogation of protein-protein interactions in live cells with a versatile ultra-high-throughput biosensor platform". Journal of Molecular Cell Biology 8 (3): 271–81. June 2016. doi:10.1093/jmcb/mjv064. PMID 26578655.
- ↑ "Target engagement and drug residence time can be observed in living cells with BRET". Nature Communications 6: 10091. December 2015. doi:10.1038/ncomms10091. PMID 26631872. Bibcode: 2015NatCo...610091R.
- ↑ "Application of BRET to monitor ligand binding to GPCRs". Nature Methods 12 (7): 661–663. July 2015. doi:10.1038/nmeth.3398. PMID 26030448.
- ↑ "NanoBRET--A Novel BRET Platform for the Analysis of Protein-Protein Interactions". ACS Chemical Biology 10 (8): 1797–804. August 2015. doi:10.1021/acschembio.5b00143. PMID 26006698.
- ↑ 40.0 40.1 "Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate". ACS Chemical Biology 7 (11): 1848–57. November 2012. doi:10.1021/cb3002478. PMID 22894855.
- ↑ 41.0 41.1 "C6-Deoxy coelenterazine analogues as an efficient substrate for glow luminescence reaction of nanoKAZ: the mutated catalytic 19 kDa component of Oplophorus luciferase". Biochemical and Biophysical Research Communications 437 (1): 23–8. July 2013. doi:10.1016/j.bbrc.2013.06.026. PMID 23792095.
- ↑ "NanoLuc product page". https://www.promega.com/products/reporter-assays-and-transfection/reporter-assays/nanoluc-luciferase-redefining-reporter-assays/.
- ↑ "Bioluminescence Profiling of NanoKAZ/NanoLuc Luciferase Using a Chemical Library of Coelenterazine Analogues". Chemistry: A European Journal 26 (4): 948–958. January 2020. doi:10.1002/chem.201904844. PMID 31765054. https://hal-pasteur.archives-ouvertes.fr/pasteur-02988525/file/Coutant_et-al_Chemistry2020-26.pdf.
- ↑ "NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells". ACS Chemical Biology 11 (2): 400–8. February 2016. doi:10.1021/acschembio.5b00753. PMID 26569370.
- ↑ "Using the novel HiBiT tag to label cell surface relaxin receptors for BRET proximity analysis". Pharmacology Research & Perspectives 7 (4): e00513. August 2019. doi:10.1002/prp2.513. PMID 31384473.
- ↑ "Homo-FRET microscopy in living cells to measure monomer-dimer transition of GFP-tagged proteins". Biophysical Journal 80 (6): 3000–8. June 2001. doi:10.1016/S0006-3495(01)76265-0. PMID 11371472. Bibcode: 2001BpJ....80.3000G.
- ↑ "Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution". Biophysical Journal 97 (9): 2613–22. November 2009. doi:10.1016/j.bpj.2009.07.059. PMID 19883605. Bibcode: 2009BpJ....97.2613B.
- ↑ Heckmeier, Philipp J.; Agam, Ganesh; Teese, Mark G.; Hoyer, Maria; Stehle, Ralf; Lamb, Don C.; Langosch, Dieter (July 2020). "Determining the Stoichiometry of Small Protein Oligomers Using Steady-State Fluorescence Anisotropy". Biophysical Journal 119 (1): 99–114. doi:10.1016/j.bpj.2020.05.025. PMID 32553128. Bibcode: 2020BpJ...119...99H.
- ↑ "Fluorescence anisotropy: from single molecules to live cells". The Analyst 135 (3): 452–9. March 2010. doi:10.1039/b920242k. PMID 20174695. Bibcode: 2010Ana...135..452G. https://zenodo.org/record/896379.
- ↑ Liu, Jiawen; Guillemeney, Lilian; Choux, Arnaud; Maître, Agnès; Abécassis, Benjamin; Coolen, Laurent (21 October 2020). "Fourier-Imaging of Single Self-Assembled CdSe Nanoplatelet Chains and Clusters Reveals out-of-Plane Dipole Contribution". ACS Photonics 7 (10): 2825–2833. doi:10.1021/acsphotonics.0c01066.
- ↑ Liu, Jiawen (April 21, 2020). "Long Range Energy Transfer in Self-Assembled Stacks of Semiconducting Nanoplatelets". Nano Letters 20 (5): 3465–3470. doi:10.1021/acs.nanolett.0c00376. PMID 32315197. Bibcode: 2020NanoL..20.3465L. https://hal.archives-ouvertes.fr/hal-02565031/file/Article%20FRET%20revised%20v1%20SOUMIS%20LE%2017%20AVRIL.pdf.
- ↑ "Resonance energy transfer: methods and applications". Analytical Biochemistry 218 (1): 1–13. April 1994. doi:10.1006/abio.1994.1134. PMID 8053542.
- ↑ Szabó, Ágnes; Szendi-Szatmári, Tímea; Szöllősi, János; Nagy, Peter (2020-07-07). "Quo vadis FRET? Förster's method in the era of superresolution". Methods and Applications in Fluorescence 8 (3): 032003. doi:10.1088/2050-6120/ab9b72. ISSN 2050-6120. PMID 32521530. Bibcode: 2020MApFl...8c2003S.
- ↑ "Using GFP in FRET-based applications". Trends in Cell Biology 9 (2): 57–60. February 1999. doi:10.1016/S0962-8924(98)01434-2. PMID 10087619.
- ↑ "Quantitative determination of the topological propensities of amyloidogenic peptides". Biophysical Chemistry 120 (1): 55–61. March 2006. doi:10.1016/j.bpc.2005.09.015. PMID 16288953.
- ↑ "Fluorescence energy transfer between ligand binding sites on aspartate transcarbamylase". Biochemistry 14 (2): 214–24. January 1975. doi:10.1021/bi00673a004. PMID 1091284.
- ↑ "Quantitative analysis of multi-protein interactions using FRET: application to the SUMO pathway". Protein Science 17 (4): 777–84. April 2008. doi:10.1110/ps.073369608. PMID 18359863.
- ↑ "The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo". Current Opinion in Structural Biology 11 (5): 573–8. October 2001. doi:10.1016/S0959-440X(00)00249-9. PMID 11785758.
- ↑ "Fluorescence resonance energy transfer analysis of cell surface receptor interactions and signaling using spectral variants of the green fluorescent protein". Cytometry 44 (4): 361–8. August 2001. doi:10.1002/1097-0320(20010801)44:4<361::AID-CYTO1128>3.0.CO;2-3. PMID 11500853.
- ↑ "A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin". Cell 102 (5): 683–94. September 2000. doi:10.1016/S0092-8674(00)00090-8. PMID 11007486.
- ↑ "Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations". The Journal of Cell Biology 160 (5): 629–33. March 2003. doi:10.1083/jcb.200210140. PMID 12615908.
- ↑ "FRET in Membrane Biophysics: An Overview". Frontiers in Physiology 2: 82. 2011-11-15. doi:10.3389/fphys.2011.00082. PMID 22110442.
- ↑ "Fluorescence-quenching and resonance energy transfer studies of lipid microdomains in model and biological membranes". Molecular Membrane Biology 23 (1): 5–16. 2006. doi:10.1080/09687860500473002. PMID 16611577.
- ↑ "Surface density determination in membranes by fluorescence energy transfer". Biochemistry 17 (24): 5241–8. November 1978. doi:10.1021/bi00617a025. PMID 728398.
- ↑ "Förster resonance energy transfer (FRET)-based small-molecule sensors and imaging agents". Chemical Society Reviews 49 (15): 5110–5139. August 2020. doi:10.1039/C9CS00318E. PMID 32697225. PMC 7408345. http://xlink.rsc.org/?DOI=C9CS00318E.
- ↑ "Dynamic visualization of cellular signaling". Nano/Micro Biotechnology. 119. Springer. 2010. 79–97. doi:10.1007/10_2008_48. ISBN 978-3-642-14946-7. Bibcode: 2010nmb..book...79N. https://books.google.com/books?id=qrGsL_wYdHMC&pg=PA79.
- ↑ "Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling". Pharmacological Reviews 64 (2): 299–336. April 2012. doi:10.1124/pr.110.004309. PMID 22407612.
- ↑ "In vivo measurement by FRET of pathway activity in bacterial chemotaxis". Two-Component Signaling Systems, Part B. Methods in Enzymology. 423. Academic Press. 2007-01-01. pp. 365–91. doi:10.1016/S0076-6879(07)23017-4. ISBN 978-0-12-373852-3.
- ↑ "Detection of caspase-3 activation in single cells by fluorescence resonance energy transfer during photodynamic therapy induced apoptosis". Cancer Letters 235 (2): 239–47. April 2006. doi:10.1016/j.canlet.2005.04.036. PMID 15958279.
- ↑ "Quantitative FRET (Förster Resonance Energy Transfer) analysis for SENP1 protease kinetics determination". Journal of Visualized Experiments (72): e4430. February 2013. doi:10.3791/4430. PMID 23463095.
- ↑ "Single-Step FRET-Based Detection of Femtomoles DNA". Sensors 19 (16): 3495. August 2019. doi:10.3390/s19163495. PMID 31405068. Bibcode: 2019Senso..19.3495S.
- ↑ "FRET-based dual-emission and pH-responsive nanocarriers for enhanced delivery of protein across intestinal epithelial cell barrier". ACS Applied Materials & Interfaces 6 (20): 18275–89. October 2014. doi:10.1021/am505441p. PMID 25260022.
- ↑ "Dual-Shell Fluorescent Nanoparticles for Self-Monitoring of pH-Responsive Molecule-Releasing in a Visualized Way". ACS Applied Materials & Interfaces 8 (29): 19084–91. July 2016. doi:10.1021/acsami.6b05872. PMID 27377369.
- ↑ "Fluorescent Peptide Dendrimers for siRNA Transfection: Tracking pH Responsive Aggregation, siRNA Binding, and Cell Penetration". Bioconjugate Chemistry 31 (6): 1671–1684. June 2020. doi:10.1021/acs.bioconjchem.0c00231. PMID 32421327. https://boris.unibe.ch/148853/1/acs.bioconjchem.0c00231.pdf.
- ↑ "Real-Time Monitoring of Nanoparticle Formation by FRET Imaging". Angewandte Chemie International Edition in English 56 (11): 2923–2926. March 2017. doi:10.1002/anie.201611288. PMID 28112478.
- ↑ "FRET-labeled siRNA probes for tracking assembly and disassembly of siRNA nanocomplexes". ACS Nano 6 (7): 6133–41. July 2012. doi:10.1021/nn3013838. PMID 22693946.
- ↑ "Application of Förster Resonance Energy Transfer (FRET) technique to elucidate intracellular and In Vivo biofate of nanomedicines". Advanced Drug Delivery Reviews. Unraveling the In Vivo Fate and Cellular Pharmacokinetics of Drug Nanocarriers 143: 177–205. March 2019. doi:10.1016/j.addr.2019.04.009. PMID 31201837.
- ↑ "Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation". Molecular Cell 9 (4): 789–98. April 2002. doi:10.1016/S1097-2765(02)00496-3. PMID 11983170.
External links
- FRET Imaging (Tutorial of Becker & Hickl, website)
 |

