Biology:Haplogroup G-M285
In human genetics, Haplogroup G-M285 or G-M342, also known as Haplogroup G1, is a Y-chromosome haplogroup. Haplogroup G1 is a primary subclade of haplogroup G.
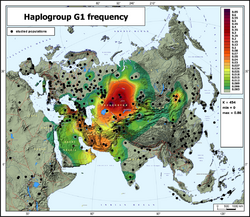
G1 is possibly believed to have originated in Iran. It has an extremely low frequency in modern populations, except (i) Iran and its western neighbors, and (ii) a region straddling south Central Siberia (Russia) and northern Kazakhstan. The most basal examples of G1 identified in living individuals, which belong to the G-L830 subclade, have been found across an area from the Arabian Peninsula (Northern Borders Region of Saudi Arabia, Ad-Dawhah of Qatar) to Ashkenazi Jews of Belarus (Minsk Region) and China (Anhui).[1][2]
Genetic features
Almost all G1 persons have the value of 12 at short tandem repeat (STR) marker DYS392 and all will have the M285 SNP mutation which characterizes this group. This value of 12 is also uncommon in other haplogroups. The M designation for M285 indicates it was first identified at Stanford University.
M285 has the following characteristics: The reference ID number is rs13447378.....Y position is 21151128....The forward primer sequence is
and the reverse sequence
....The mutation is G to C.[3] M285 was first reported in 2004 by Cinnioglu et al.[4] So far all persons tested who have the M285 mutation also are positive the M342 SNP mutation. If someone should test differently, his results would become the basis of a new G1 category. M342 is located at sequence position 21653330 and is
in the forward primer and
in the reverse. It is a mutation from C to T.[4] M342 was also identified at Stanford University,
In addition, within haplogroup G, those G1 persons tested so far are uniquely positive (derived) for SNP L89 while all other G persons are negative (ancestral). The L89 SNP mutation has also been found in other haplogroups. L89 is located at sequence position 8038725, and the reference ID number is rs35160044. It is a mutation from T to C. The L89 designation was provided by Family Tree DNA.
Dating of G1 origin
While research studies have not yet dated the origin of G1's M285 SNP mutation, it seemingly represents one of the older G groups, arising perhaps halfway between the origin of G and the present day based on the number of STR marker mutations.
Geographical distribution
Geographical distribution
While most research studies have failed to test for G1, likely or proven examples have been found in four groups (by geography and/or and/or secondary subclade):
- Iran/Central Asia (including Russian Central Siberia, Kazakhstan and Pakistan );
- the Middle East/South-East Europe (e.g. Greece, Turkey, Iraq, Dubai, Palestine, Jordan, Saudi Arabia and Yemen);
- in very small numbers in South Asia (e.g. Nepal, India and Sri Lanka), and;
- at very low frequencies in the rest of Europe.[5]
The highest reported concentration of G1 and its subgroups in a single country is in Iran, with next most frequent concentrations in neighboring countries to the west. Specifically G1 was found in 5% of 177 samples from southern Iran in one study. The same study found G1 in 3% of 33 samples in northern Iran. The percentage in the south was almost equal to the percentage of G2, the only region in the world where G2 types were not found to be heavily dominant.[6] Earlier studies which found about the same percentages of G in various parts of Iran failed to test specifically for G1.[7][8]
In Turkey, Cinnioglu found 1% of 523 samples of all types belonging to haplogroup G1 and like all available G1 outside Iran G1 was overshadowed by a much larger number of G2 men in the sample. All but one of the G1 samples in this study belonged to the further subgroup G1a, and all the samples were from northeastern Turkey.[9]
A study of Lebanon by Zalloua et al. failed to test for G1, but 26% of 38 G samples had the value of 12 for STR marker DYS392, almost always characteristic of G1. These G1 samples represent only 2% of the 587 total Lebanese samples and were well distributed among all religious groups there.[10]
A broader study of the Levant by El-Sibai et al. also failed to test for G1, but 12% of 17 Syrian G samples had the 12 value at DYS392. These G1 samples represent less than 1% of 354 Syrian samples. In the same study, 21% of 14 Jordanian G samples had the 12 value. These G1 samples represent 1% of 274 Jordanian samples. None of 8 Egyptian G samples had the 12 value.[11]
The likely G samples in the YHRD database [12] from the southern Caucasus area (Azerbaijan, Georgia, Armenia, Abazinia, Abkhazia) and from the northern Caucasus (North Ossetians, Kabardinians, Ingushians, Darginians, Lezginians, Rutulians and Chechnians) have only several 12-value DYS392 samples that might be G1.
Sites with G1 concentrations
The highest concentration of G1 within a distinct group within a country was reported among the kazakh tribe Argyn - 67% and its branch Madjars of Kazakhstan where G1 persons comprised 87% of 45 samples,.[13]
G1 subgroups
SNP-defined subgroups
Subgroup G1a (P20) is rather common among G1 persons, but the reliability of SNP P20 in identifying G1a persons makes some tests for P20 a problem.[14] P20 results may be reported as P20.1, P20.2 and P20.3, and persons may have varying results for each. The technical features of P20 are: Y position is 25029911; 23396163.....forward primer is
.....reverse primer is
...the mutation is a deletion of C.[3] The mutation was identified at the University of Arizona and first reported by the Y Chromosome Consortium in 2002.[15] Category G1a has a separate subgroup based on the presence of the value of 8 at short tandem repeat marker DYS494. With the exception of this G1a subgroup, all other G persons have 9 at this slowly mutating marker. So far about half of G1a persons who are otherwise ungrouped have this 8 value.[16]
G1a1 (L201, L202 and L203) were identified in a G1a person tested at Family Tree DNA in fall 2009, Subsequent testing showed that only some G1a persons have these mutations, and so far they are all from a closely related Ashkenazi Jewish cluster of men. The following Y positions were noted: 2718285 for L201, 13001714 for L202 and 13001715 for L203. In L201 the mutation is C to T; in L202 from T to C; and in L203 from A to G. If any man should have one of these SNP mutations without the others, this information would become the basis of a new G1a1 classification.
Old G1b (P76) was removed from the official listing in August, 2012, because its discovery in a single person makes it so far only a private SNP.
G1b (L830, L831, L832, L834, L835) were identified in late 2011 at Family Tree DNA. The G1c mutations are: L830 (T to C at position 6923908), L831 (T to C at position 6991562), L832 (T to G at position 12796626), L834 (T to A at position 16019950), L835 (G to A at position 16291409).
STR marker value clusters
There exist two distinctive European-ancestry Ashkenazi Jewish clusterings within G1a1 and G1c and a distinctive G1a Kazakh cluster—all three based on short tandem repeat (STR) marker values.[17] Men in the Jewish G1c cluster have a value of 12 at STR marker DYS446 which is several values lower than almost all G persons, but the other two groups lack one specific marker value as an identifier. When approximately 30 or more STR markers are available when comparing samples, members in each of the three groups are easily ascertained. One of the distinctive European-ancestry G1c Ashkenazi Jewish STR marker value combinations is found also in an Iraqi Jew in one research study.[18] See also page covering Jews with Haplogroup G (Y-DNA).
Among available 67-marker G1 STR samples,[17] the Ashkenazi G1a1 Jewish cluster represents the closest relatives to the Kazakh G1a cluster based on similarity of STR marker values.
The Ashkenazi G1c cluster is only distantly related to the Ashkenazi G1a1 cluster, and the G1c Ashkenazi cluster has nearest relatives instead among diverse Europeans.
Migrations of G1 persons
The concentration of G1 found today in Iran and adjoining territory to its west suggests, but does not prove, this same area was the site of G1 origin. Due to the lengthy age of G1, distant migrations of small numbers of G1 persons could have occurred at any time during the historical period through a variety of types of population movements. It would be speculative to assume how the Ashkenazi G1 clusters or the Kazakh G1 clusters of men arrived in northeastern Europe and Kazakhstan respectively.
See also
- genetic genealogy
- Y-DNA haplogroups by ethnic groups
- Haplogroup G (Y-DNA) Country by Country
References
- ↑ Jump up to: 1.0 1.1 YFull YTree v8.07.00 accessed on August 30, 2020
- ↑ "Welcome to FamilyTreeDNA Discover (Beta)" (in en). https://discover.familytreedna.com/y-dna/G-BY21343/projects.
- ↑ Jump up to: 3.0 3.1 Karafat, T. (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–38. doi:10.1101/gr.7172008. PMID 18385274.
- ↑ Jump up to: 4.0 4.1 Cinnioglu, C. (2004). "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics 114 (2): 127–48. doi:10.1007/s00439-003-1031-4. PMID 14586639. Archived from the original on 2006-06-19. https://web.archive.org/web/20060619105314/http://hpgl.stanford.edu/publications/HG_2004_v114_p127-148.pdf.
- ↑ |url=https://sites.google.com/site/haplogroupgproject/project-roster Haplogroup G Project samples
- ↑ "Iran: tricontinental nexus for Y-chromosome driven migration". Human Heredity 61 (3): 132–43. 2006. doi:10.1159/000093774. PMID 16770078. http://content.karger.com/produktedb/produkte.asp?typ=fulltext&file=HHE2006061003132.
- ↑ Nasidze, I (2006). "Concomitant Replacement of Language and mtDNA in South Caspian Populations of Iran". Current Biology 16 (7): 668–73. doi:10.1016/j.cub.2006.02.021. PMID 16581511.
- ↑ Nasidze, I (2008). "Close Genetic Relationship Between Semitic-speaking and Indo-European-speaking Groups in Iran". Annals of Human Genetics 72 (Pt 2): 241–52. doi:10.1111/j.1469-1809.2007.00413.x. PMID 18205892.
- ↑ Cinnioglu, C. (2004). "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics 114 (2): 127–48. doi:10.1007/s00439-003-1031-4. PMID 14586639.
- ↑ Zalloua, P. (2008). "Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean". American Journal of Human Genetics 83 (5): 633–42. doi:10.1016/j.ajhg.2008.10.012. PMID 18976729.
- ↑ El-Sibai, M. (2009). "Geographical structure of the Y-chromosomal genetic landscape of the Levant: a coastal-inland contrast". Annals of Human Genetics 73 (6): 568–81. doi:10.1111/j.1469-1809.2009.00538.x. PMID 19686289.
- ↑ |url=http://www.yhrd.org/Search
- ↑ Biro, A. et al. (2009). "A Y-Chromosomal Comparison of the Madjars (Kazakhstan) and the Magyars (Hungary)". American Journal of Physical Anthropology 139 (3): 305–10. doi:10.1002/ajpa.20984. PMID 19170200. http://www3.interscience.wiley.com/journal/121660348/abstract.
- ↑ http://archiver.rootsweb.ancestry.com/th/read/GENEALOGY-DNA/2007-07/1185576938 Report of Thomas Krahn
- ↑ The Y Chromosome Consortium (2002). "A nomenclature system for the tree of human Y-chromosomal binary haplogroups". Genome Research 12 (2): 339–48. doi:10.1101/gr.217602. PMID 11827954.
- ↑ Haplogroup G Project results |url=https://sites.google.com/site/haplogroupgproject/nonstandard-markers
- ↑ Jump up to: 17.0 17.1 |url=https://sites.google.com/site/haplogroupgproject/project-roster
- ↑ Hammer, M. (2009). "Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood". Human Genetics 126 (5): 719–24. doi:10.1007/s00439-009-0727-5. PMID 19669163.
External links
- Haplogroup G Project Site
- Spread of Haplogroup G, from National Geographic
- Haplogroup G tutorial from Genebase
- Y-DNA Haplogroup G and its subclades from the 2010 ISOGG haplotree
- Balanovsky, Oleg; Zhabagin, Maxat; Agdzhoyan, Anastasiya; Chukhryaeva, Marina; Zaporozhchenko, Valery; Utevska, Olga; Highnam, Gareth; Sabitov, Zhaxylyk et al. (2015). "Deep Phylogenetic Analysis of Haplogroup G1 Provides Estimates of SNP and STR Mutation Rates on the Human Y-Chromosome and Reveals Migrations of Iranic Speakers". PLOS ONE 10 (4): e0122968. doi:10.1371/journal.pone.0122968. PMID 25849548. Bibcode: 2015PLoSO..1022968B.
 |