Biology:Haplogroup J-M172
| Haplogroup J-M172 | |
|---|---|
 | |
| Possible time of origin | 32000 ybp[1] |
| Coalescence age | 28000 ybp[1] |
| Possible place of origin | Upper Mesopotamia, Western Iran[2] |
| Ancestor | J-P209 |
| Defining mutations | M172 |
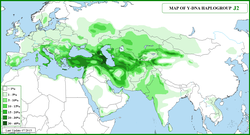
| Highest frequencies | Ingush 88.8% (Balanovsky 2011), Chechens 56.7% (Balanovsky 2011), Georgians 21% (Wells 2001)-72%, Azeris 24% (Di Giacomo 2004)-48% (Wells 2001), Iraqis 24%(Al-Zahery 2011)-25% Al-Zahery 2003 and Sanchez 2005, Cretans 35% (El-Sibai 2009), Uyghurs 34% (Shou 2010),[3] Yaghnobis 32% (Wells 2001), Uzbeks 30.4% (Shou 2010), Greeks 10%-48%(Martinez 2007), Muslim Kurds 28.4%(Nebel 2001), Lebanese 30% (Semino 2004) [dubious ](Wells 2001), Ashkenazi Jews 24-30%(Nebel 2001) (Semino 2004), Turks 24% (Cinnioglu 2004)-40% (Semino 2000), Hazara 26.6% (Haber et al, 2012),[4] Kuwaiti 26% [citation needed] and (Wells 2001), Cypriots 12.9% (El-Sibai 2009)-37% (Capelli 2005),[5] Abkhaz 25% (Nasidze 2004), Iranians 22.5%(Grugni 2012)-24%,[6] Balkars 24% (Battaglia 2008), Italians 9%-36%(Capelli 2007) and (Semino 2000), Armenians 21%(Wells 2001)-24% (Nasidze 2004), Palestinians 29%(Nebel 2001)[citation needed], Mordvins 15.3%,[7] Kazan Tatars 15.1%,[7] Chuvash 14%,[7] Sephardi Jews 15 -20% (Shen 2004)-29% (Nebel 2001), Ossetians 16%(Balanovsky 2011)-24%(Nasidze 2004), Circassians 21.8% (Balanovsky 2011), Maltese 21% (Capelli 2005), Lemba 20.8% (Soodyall 2013), North Indian Shia Muslims 18% (Eaaswarkhanth 2009), Albanians 16% (Battaglia 2008), Syrians 14% (Di Giacomo 2004)-29%[citation needed], and Kalash people 9.1%.(Firasat 2007)[8] |
In human genetics, Haplogroup J-M172 or J2[Phylogenetics 1] is a Y-chromosome haplogroup which is a subclade (branch) of haplogroup J-M304.[Phylogenetics 2] Haplogroup J-M172 is common in modern populations in Western Asia, Central Asia, South Asia, Southern Europe, Northwestern Iran and North Africa. It is thought that J-M172 may have originated between the Caucasus, Anatolia and/or Western Iran.[9][10]
It is further divided into two complementary clades, J-M410 and J-M12 (M12, M102, M221, M314).
Origins
The date of origin for haplogroup J-M172 was estimated by Batini et al in 2015 as between 19,000 and 24,000 years before present (BP).[11] Samino et al in 2004 dated the origin of the parent haplogroup, J-P209, to between 18,900 and 44,500 YBP.[12] Ancient J-M410, specifically subclade J-Y12379*, has been found, in a mesolithic context, in a tooth from the Kotias Klde Cave in western Georgia dating 9.529-9.895 cal. BP.[13] This sample has been assigned to the Caucasus hunter-gatherers (CHG) autosomal component.[14] J-M410, more specifically its subclade J-PF5008, has also been found in a mesolithic sample from the Hotu and Kamarband Caves located in Mazandaran Province of Iran, dating back to 9,100-8,600 B.C.E (approximately 11,000 ybp).[15] Both samples belong to the Trialetian Culture. It is likely that J2 men had settled over most of Anatolia, the South Caucasus and the Zagros mountains by the end of the Last Glaciation 12,000 years ago.[16]
Zalloua and Wells 2004 and al-Zaheri 2003 claimed to have uncovered the earliest known migration of J2, expanded possibly from Anatolia and the Caucasus.[9][10][17] In 2001, Nebel et al. found that, "According to Underhill et al. (2000), Eu 9 (H58) evolved from Eu 10 (H71) through a T→G transversion at M172 (emphasis added)," and that in today's populations, Eu 9 (the post-mutation form of M172) is strongest in the Caucasus, Asia Minor and the Levant, whilst Eu 10 becomes stronger and replaces the frequency of Eu 9 as one moves south into the Arabian Peninsula,[18] so that people from the Caucasus met with Arabs near and between Mesopotamia (Sumer/Assyria) and the Negev Desert, as "Arabisation" spread from Arabia to the Fertile Crescent and Turkey.
Per research by Di Giacomo 2004, J-M172 haplogroup spread into Southern Europe from either the Levant or Anatolia, likely parallel to the development of agriculture.[19] As to the timing of its spread into Europe, Di Giacomo points to events which post-date the Neolithic, in particular the demographic floruit associated with the rise of the Ancient Greek world. Semino et al. derived older age estimates for overall J2 (having used the Zhivotovsky method c.f. Di Giacomo), postulating its initial spread with Neolithic farmers from the Near East. However, its subclade distribution, showing localized peaks in the Southern Balkans, southern Italy, north/central Italy and the Caucasus, does not conform to a single 'wave-of-advance' scenario, betraying a number of still poorly understood post-Neolithic processes which created its current pattern. Like Di Giacomo, the Bronze Age southern Balkans was suggested by Semino 2004 to have been an important vector of spread.[12]
Distribution
Haplogroup J-M172 is found mainly in the Fertile Crescent, the Caucasus (Nasidze 2003), Anatolia, Italy, the Mediterranean littoral, and the Iranian plateau (Semino 2004). Y-DNA: J2 (J-M172): Syrid/Nahrainid Arabid(s).
The highest reported frequency of J-M172 ever was 87.4%, among Ingush in Malgobek (Balanovsky 2011).
More specifically it is found in Iraq (Al-Zahery 2003), Kuwait,[20] Syria (Luis 2004), Lebanon (Zalloua 2008l), Turkey (Cinnioglu 2004), Georgia (Nasidze 2003), Azerbaijan (Di Giacomo 2004), North Caucasus (Nasidze 2004), Armenia (Wells 2001), Iran (Nasidze 2004), Israel (Semino 2004), Palestine (Semino 2004), Cyprus (Capelli 2005), Greece (Martinez 2007), Albania (Semino 2000), Italy (Capelli 2007), Spain (Di Giacomo 2003), and more frequently in Iraqis 24% (Al-Zahery 2011), Chechens 51.0%-58.0% (Balanovsky 2011), Georgians 21% (Wells 2001)-72% (Wells 2001), Lebanese 30% (Semino 2004), Ossetians 24% (Nasidze 2004), Balkars 24% (Battaglia 2008), Syrians 23% (Luis 2004), Turks 13% (Cinnioglu 2004)-40% (Semino 2000), Cypriots 12.9% (El-Sibai 2009)-37% (Capelli 2005), Armenians 21% (Wells 2001)-24% (Nasidze 2004), Circassians 21.8%(Balanovsky 2011), Iranians 10% (Nasidze 2004)-25% (Wells 2001), Albanians 16% (Battaglia 2008) and (Semino 2000), Italians 9%-36% (Capelli 2007), Sephardi Jews 15% (Nebel 2001)-29%(Semino 2004), Maltese 21% (Capelli 2005), Palestinians 17% (Semino 2004), Saudis 14% (Abu-Amero 2009), Jordanians 14%, Omanis 10%-15% (Di Giacomo 2004) and (Luis 2004) and North Indian Shia Muslim 18% (Eaaswarkhanth 2009).
North Africa
Haplogroup J2 is found with low frequencies in North Africa. with a hotspot in Sousse region Fadhlaoui-Zid 2014 most of Sousse samples have the same haplotypes found in Haplogroup J-L271 which was found in Msaken.
| Country/Region | Sampling | N | J-M172 | Study |
| Tunisia | Tunisia | 62 | 8 | El-Sibai 2009 |
| Tunisia | Sousse | 220 | 8.2 | Fadhlaoui-Zid 2014 |
| Algeria | Oran | 102 | 4.9 | Robino 2008 |
| Egypt | 124 | 7.6 | El-Sibai 2009 | |
| Egypt | 147 | 12.0 | Abu-Amero 2009 | |
| Morocco | 221 | 4.1 | Fregel 2009 | |
| North Africa | Algeria, Tunisia | 202 | 3.5 | Fregel 2009 |
Central Asia
| Country/Region | Sampling | N | J-M172 | Study |
| Xinjiang | Lop Uyghurs | 64 | 57.8 | Liu 2018 |
| Xinjiang | Uyghurs | 50 | 34 | Shou 2010 |
| Tajikistan | Yaghnobis | 31 | 32 | Wells 2001 |
| Dushanbe | Tajiks | 16 | 31 | Wells 2001 |
| Xinjiang | Uzbeks | 23 | 30.4 | Shou 2010 |
| Afghanistan | Hazara | 60 | 26.6 | Haber 2012 |
| Xinjiang | Keriyan Uyghurs | 39 | 25.6 | Liu 2018 |
| Kazakhstan | Uyghurs | 41 | 20 | Wells 2001 |
| Samarkand | Tajiks | 40 | 20 | Wells 2001 |
| Tajikistan | Tajiks | 38 | 18.4 | Wells 2001 |
| Turkmenistan | Turkmens | 30 | 17 | Wells 2001 |
| Xinjiang | Pamiri Tajiks | 31 | 16.1 | Shou 2010 |
| Afghanistan | Uzbeks | 126 | 16 | Di Cristofaro 2013 |
| Bukhara | Uzbeks | 58 | 16 | Wells 2001 |
| Samarkand | Uzbeks | 45 | 16 | Wells 2001 |
| Surkhandarya | Uzbeks | 68 | 16 | Wells 2001 |
| Uzbekistan | Uzbeks | 366 | 13.4 | Wells 2001 |
| Kazakhstan | Kazakhs | 30 | 13.3 | Karafet 2001 |
| Turpan area | Uyghurs | 143 | 9.8 | [citation needed] |
| Hotan area | Uyghurs | 478 | 9.2 | [citation needed] |
| Changji | Hui | 175 | 9.1 | [citation needed] |
| Xinjiang | Dolan Uyghurs | 76 | 7.9 | Liu 2018 |
| Ningxia | Hui | 65 | 7.7 | [citation needed] |
| Kizilsu | Kyrgyz | 241 | 6.64% | Guo 2020 |
| Kazakhstan | Kazakhs | 1294 | 4.33% | Ashirbekov 2017 |
| Kyrgyzstan | Kyrgyz | 132 | 3.79% | Di Cristofaro 2013 |
J-M172 is found at moderate frequencies among Central Asian people such as Uyghurs, Uzbeks, Turkmens, Tajiks, Kazakhs, and Yaghnobis. According to the genetic study in Northwest China by Shou et al. (2010), a notable high frequency of J-M172 is observed particularly in Uyghurs 34% and Uzbeks 30.4% in Xinjiang, China . Liu Shuhu et al. (2018) found J2a1 (L26/Page55/PF5110/S57, L27/PF5111/S396) in 43.75% (28/64) and J2a2 (L581/S398) in 14.06% (9/64) of a sample of Lop Uyghurs from Qarchugha Village of Yuli (Lopnur) County, Xinjiang, J2a1b1 (M92, M260/Page14) in 25.64% (10/39) of a sample of Keriyan Uyghurs from Darya Boyi Village of Yutian (Keriya) County, Xinjiang, and J2a1 (L26/Page55/PF5110/S57, L27/PF5111/S396) in 3.95% (3/76) and J2a2 (L581/S398) in 3.95% (3/76) of a sample of Dolan Uyghurs from Horiqol Township of Awat County, Xinjiang.[21] Only far northwestern ethnic minorities had haplogroup J in Xinjiang, China. Uzbeks in the sample had 30.4% J2-M172 and Tajiks of Xinjiang and Uyghurs also had it.[22]
The haplogroup has an ancient presence in Central Asia and seems to have preceded the spread of Islam (Shou 2010). In addition, the immediate ancestor of J-M172, namely J* (J-M304*, a.k.a. J-P209*, J-12f2.1*) is also found among Xibo, Kazakh, Dongxiang and Uzbek people in Northwest China.
In 2015, two ancient samples belonging to J-M172 or J-M410 (J2a) were found at two different archaeological sites in Altai, eastern Russia : Kytmanovo and Sary-bel kurgan. Both of the ancient samples are related to Iron Age cultures in Altai. Sary-bel J2/J2a is dated to 50 BC whereas Kytmanovo sample is dated to 721-889 AD. Genetic admixture analysis of these samples also suggests that the individuals were more closely related to West Eurasians than other Altaians from the same period, although they also seem to be related to present-day Turkic peoples of the region.[23][24][25]
Europe
| Country/Region | Sampling | N | J-M172 | Study |
| Albania | 55 | 19.9% 11/55 |
Battaglia 2009 | |
| Bosnia-Herzegovina | Serbs | 81 | 8.7 | Battaglia 2009 |
| Cyprus | 164 | 12.9 | El-Sibai 2009 | |
| Greece | Crete | 143 | 35 | El-Sibai 2009 |
| Iberia | 655 | 7 | Fregel 2009 | |
| Iberia | 1140 | 7.7 | Adams 2008 | |
| Italy | Sicily | 212 | 22.6 | El-Sibai 2009 |
| Italy | Mainland | 699 | 20 | Capelli 2007 |
| Italy | Central Marche | 59 | 35.6 | Capelli 2007 |
| Italy | West Calabria | 57 | 35.1 | Capelli 2007 |
| Italy | Val Badia | 34 | 8.8 | Capelli 2007 |
| Malta | 90 | 21.1 | El-Sibai 2009 | |
| Portugal | North, Center, South | 303 | 6.9 | El-Sibai 2009 |
| Portugal | Tras-os-Montes (Jews) | 57 | 24.5 | Nogueiro 2010 |
| Sardinia | 81 | 9.9 | El-Sibai 2009 | |
| Spain | Mallorca | 62 | 8.1 | El-Sibai 2009 |
| Spain | Sevilla | 155 | 7.8 | El-Sibai 2009 |
| Spain | Leon | 60 | 5 | El-Sibai 2009 |
| Spain | Ibiza | 54 | 3.7 | El-Sibai 2009 |
| Spain | Cantabria | 70 | 2.9 | El-Sibai 2009 |
| Spain | Galicia | 292 | 13 | [citation needed] |
| Spain | Canary Islands | 652 | 10.5 | Fregel 2009 |
In Europe, the frequency of Haplogroup J-M172 drops as one moves northward away from the Mediterranean. In Italy, J-M172 is found with regional frequencies ranging between 9% and 36% (Capelli 2007). In Greece, it is found with regional frequencies ranging between 10% and 48%. Approximately 24% of Turkish men are J-M172 according to a recent study, (Cinnioglu 2004) with regional frequencies ranging between 13% and 40% (Semino 2000). Combined with J-M267, up to half of the Turkish population belongs to Haplogroup J-P209.
It has been proposed that haplogroup subclade J-M410 was linked to populations on ancient Crete by examining the relationship between Anatolian, Cretan, and Greek populations from around early Neolithic sites in Crete. Haplogroup J-M172 was associated with Neolithic Greece (ca. 8500 - 4300 BCE) and was reported to be found in modern Crete (3.1%) and mainland Greece (Macedonia 7.0%, Thessaly 8.8%, Argolis 1.8%) (King 2008).
North Caucasus
| Country/Region | Sampling | N | J-M172 | Study |
| Caucasus | Abkhaz | 58 | 13.8 | Balanovsky 2011 |
| Caucasus | Avar | 115 | 6 | Balanovsky 2011 |
| Caucasus | Chechen | 330 | 57 | Balanovsky 2011 |
| Caucasus | Adyghe | 142 | 21.8 | Balanovsky 2011 |
| Caucasus | Dargins | 101 | 1 | Balanovsky 2011 |
| Caucasus | Ingush | 143 | 88.8 | Balanovsky 2011 |
| Caucasus | Kaitak | 33 | 3 | Balanovsky 2011 |
| Caucasus | Kumyks | 73 | 21 | Yunusbayev 2012 |
| Caucasus | Kubachi | 65 | 0 | Balanovsky 2011 |
| Caucasus | Lezghins | 81 | 2.5 | Balanovsky 2011 |
| Caucasus | Ossets | 357 | 16 | Balanovsky 2011 |
| Caucasus | Shapsug | 100 | 6 | Balanovsky 2011 |
| Caucasus | 1525 | 28.1 | Balanovsky 2011 |
J-M172 is found at very high frequencies in certain peoples of the Caucasus: among the Ingush 87.4% (Balanovsky 2011), Chechens 55.2% (Balanovsky 2011), Georgians 21%-72%, (Wells 2001), Azeris 24% (Di Giacomo 2004)-48%, (Wells 2001) Abkhaz 25%, (Nasidze 2004) Balkars 24% (Battaglia 2008), Ossetians 24% (Nasidze 2004), Armenians 21% (Wells 2001)-24% (Nasidze 2004), Adyghe 21.8% (Balanovsky 2011), and other groups ( Nasidze 2004 and Nasidze 2003).
West Asia
| Country/Region | Sampling | N | J-M172 | Study |
| Jewish | Ashkenazim Jewish | 442 | 19 | Behar 2004 |
| Iran | 92 | 25 | El-Sibai 2009 | |
| Iraq | 154 | 24 | Al-Zahery 2011[26] | |
| Palestinian Arab | Akka | 101 | 18.6 | El-Sibai 2009 |
| Jordan | 273 | 14.6 | El-Sibai 2009 | |
| Lebanon | 951 | 29.4 | El-Sibai 2009 | |
| Oman | 121 | 10.0 | Abu-Amero 2009 | |
| Qatar | 72 | 8.3 | El-Sibai 2009 | |
| Saudi Arabia | 157 | 14 | Abu-Amero 2009[27] | |
| Syria | Syria | 554 | 20.8 | El-Sibai 2009 |
| Turkey | 523 | 24.2 | El-Sibai 2009 | |
| UAE | 164 | 10.3 | El-Sibai 2009 | |
| Yemen | 62 | 9.6 | El-Sibai 2009 |
Sephardi Jews have about 15% (Nebel 2001)-29% (Semino 2004), of haplogroup J-M172, and Ashkenazi Jews have 15% (Shen 2004)-23% (Semino 2004). It was reported in an early study which tested only four STR markers (Malaspina 2001) that a small sample of Italian Cohens belonged to Network 1.2, an early designation for the overall clade now known as J-L26, defined by the deletion at DYS413. However, a large number of all Jewish Cohens in the world belong to haplogroup J-M267 (see Cohen modal haplotype).
Haplogroup J-M172 has been shown to have a more northern distribution in the Middle East, although it exists in significant amounts in the southern middle-east regions, a lesser amount of it was found when compared to its brother haplogroup, J-M267, which has a high frequency southerly distribution. It was believed that the source population of J-M172 originated from the Levant/Syria (Syrid-J-M172), and that its occurrence among modern populations of Europe, Central Asia, and South Asia was a sign of the neolithic agriculturalists. However, as stated it is now believed more likely to have been spread in waves, as a result of post-Neolithic processes .
South Asia
Haplogroup J2 has been present in South Asia mostly as J2a-M410 and J2b-M102, since neolithic times (9500 YBP).[28][29] J2-M172 was found to be significantly higher among Dravidian castes at 19% than among Indo-Aryan castes at 11%. J2-M172 and J-M410 is found 21% among Dravidian middle castes, followed by upper castes, 18.6%, and lower castes 14%.[30] Among caste groups, the highest frequency of J2-M172 was observed among Tamil Vellalars of South India, at 38.7%.[30] J2 is present in Indian tribals too and has a frequency of 11% in Austro-Asiatic tribals. Among the Austro-Asiatic tribals, the predominant J2 occurs in the Asur tribe (77.5%) albeit with a sample size of 40[28] and in the Lodha (35%) of West Bengal.[30] J2 is also present in the South Indian hill tribe Toda at 38.46% albeit with a sample size of only 26,[31] in the Andh tribe of Telangana at 35.19%,[32] in the Narikuravar tribe at 57.9%[28] and in the Kol tribe of Uttar Pradesh at a frequency of 33.34%.[33] Haplogroup J-P209 was found to be more common in India's Shia Muslims, of which 28.7% belong to haplogroup J, with 13.7% in J-M410, 10.6% in J-M267 and 4.4% in J2b (Eaaswarkhanth 2009).
In Pakistan , the highest frequencies of J2-M172 were observed among the Parsis at 38.89%, the Dravidian speaking Brahui's at 28.18% and the Makrani Balochs at 24%.[34] It also occurs at 18.18% in Makrani Siddis and at 3% in Karnataka Siddis.[34][35]
J2-M172 is found at an overall frequency of 16.1% in the people of Sri Lanka.[36] In Maldives, 22% of Maldivian population were found to be haplogroup J2 positive.[37] Subclades of M172 such as M67 and M92 were not found in either Indian or Pakistani samples which also might hint at a partial common origin.[30]
J2-M172 has been observed in 15.9% (20/164 J2a-M410, 6/164 J2b2-M241) of Tharu from Uttar Pradesh,[38] 13.4% (19/202 J2a-M410, 8/202 J2b2-M241) of Tharu from Nepal,[39][38] and 8.9% (4/45 J2a-M410) of Tharu from Uttarakhand.[38]
Subclade distribution
Haplogroup J-M172 is subdivided into two complementary sub-haplogroups: J-M410, defined by the M410 genetic marker, and J-M12, defined by the M12 genetic marker.
J-M172
J-M172 is typical of populations of the Near East, Southern Europe, Southwest Asia and the Caucasus, with a moderate distribution through much of Central Asia, South Asia, and North Africa.[40]
J-M410
J-M410* is found in Georgia, North Ossetia.[41]
J-M47
J-M47 is found with low frequency in Georgia, (Battaglia 2008) southern Iran (Regueiro 2006), Qatar (Cadenas 2008) Saudi Arabia (Abu-Amero 2009), Syria (Di Giacomo 2004), Tunisia (Arredi 2004), Turkey (Di Giacomo 2004 and Cinnioglu 2004), the UAE, (Cadenas 2008), and Central Asia/Siberia (Underhill 2000).
J-M67
J-M67 (called J2f in older papers) has its highest frequencies associated with Nakh peoples. Found at very high (majority) frequencies among Ingush in Malgobek (87.4%), Chechens in Dagestan (58%), Chechens in Chechnya (56.8%) and Chechens in Malgobek, Ingushetia (50.9%) (Balanovsky 2011). In the Caucasus, it is found at significant frequencies among Georgians (13.3%) (Semino 2004), Iron Ossetes (11.3%), South Caucasian Balkars (6.3%) (Semino 2004), Digor Ossetes (5.5%), Abkhaz (6.9%), and Cherkess (5.6%) (Balanovsky 2011). It is also found at notable frequencies in the Mediterranean and Middle East, including Cretans (10.2%), North-central Italians (9.6%), Southern Italians (4.2%; only 0.8% among N. Italians), Anatolian Turks (2.7-5.4%), Greeks (4-4.3%), Albanians (3.6%), Ashkenazi Jews (4.9%), Sephardis (2.4%), Catalans (3.9%), Andalusians (3.2%), Calabrians (3.3%), Albanian Calabrians (8.9%) (see Di Giacomo 2004 and Semino 2004).
J-M92/M260, a subclade of J-M67, has been observed in 25.64% (10/39) of a sample of Keriyan Uyghurs from Darya Boyi Village of Yutian (Keriya) County, Xinjiang.[21] This Uyghur village is located in a remote oasis in the Taklamakan Desert.
J-M319
J-M319 is found with low to moderate frequency in Cretan Greeks (Martinez 2007 and King 2008), Iraqi Jews (Shen 2004), and Moroccan Jews (Shen 2004).
J-M158
J-M158 (location under L24 uncertain) J-M158 is found with low frequency in Turkey (Cinnioglu 2004), South Asia (Sengupta 2006 and Underhill 2000), Indochina (Underhill 2000), and Iberian Peninsula.
Phylogenetics
In Y-chromosome phylogenetics, subclades are the branches of haplogroups. These subclades are also defined by single nucleotide polymorphisms (SNPs) or unique event polymorphisms (UEPs).
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| J-12f2a | 9 | VI | Med | 23 | Eu10 | H4 | B | J* | J | J | J | - | - | - | - | - | - | J |
| J-M62 | 9 | VI | Med | 23 | Eu10 | H4 | B | J1 | J1a | J1a | J1a | - | - | - | - | - | - | Private |
| J-M172 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2* | J2 | J2 | J2 | - | - | - | - | - | - | J2 |
| J-M47 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2a | J2a | J2a1 | J2a4a | - | - | - | - | - | - | J2a1a |
| J-M68 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2b | J2b | J2a3 | J2a4c | - | - | - | - | - | - | J2a1c |
| J-M137 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2c | J2c | J2a4 | J2a4h2a1 | - | - | - | - | - | - | J2a1h2a1a |
| J-M158 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2d | J2d | J2a5 | J2a4h1 | - | - | - | - | - | - | J2a1h1 |
| J-M12 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2e* | J2e | J2b | J2b | - | - | - | - | - | - | J2b |
| J-M102 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2e1* | J2e1 | J2b | J2b | - | - | - | - | - | - | J2b |
| J-M99 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2e1a | J2e1a | J2b2a | J2b2a | - | - | - | - | - | - | Private |
| J-M67 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2f* | J2f | J2a2 | J2a4b | - | - | - | - | - | - | J2a1b |
| J-M92 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2f1 | J2f1 | J2a2a | J2a4b1 | - | - | - | - | - | - | J2a1b1 |
| J-M163 | 9 | VI | Med | 24 | Eu9 | H4 | B | J2f2 | J2f2 | J2a2b | J2a4b2 | - | - | - | - | - | - | Private |
Research publications
The following research teams per their publications were represented in the creation of the YCC Tree.
Phylogenetic trees
There are several confirmed and proposed phylogenetic trees available for haplogroup J-M172. The scientifically accepted one is the Y-Chromosome Consortium (YCC) one published in Karafet 2008 and subsequently updated. A draft tree that shows emerging science is provided by Thomas Krahn at the Genomic Research Center in Houston, Texas. The International Society of Genetic Genealogy (ISOGG) also provides an amateur tree.[Phylogenetics 3][42]
The Genomic Research Center draft tree
This is Thomas Krahn at the Genomic Research Center's draft tree Proposed Tree for haplogroup J-M172 (Krahn FTDNA). For brevity, only the first three levels of subclades are shown.
- M172, L228
- M410, L152, L212, L505, L532, L559
- PF5008
- Y182822
- L581
- Z37823
- L581
- Y182822
- PF4610
- Z6046
- L26
- PF5008
- M12, M102, M221, M314, L282
- M205
- M241
- M99
- M280
- M321
- P84
- L283
- M410, L152, L212, L505, L532, L559
The Y-Chromosome Consortium tree
This is the official scientific tree produced by the Y-Chromosome Consortium (YCC). The last major update was in 2008 (Karafet 2008). Subsequent updates have been quarterly and biannual. The current version is a revision of the 2010 update.[43]
The ISOGG tree
Below are the subclades of Haplogroup J-M172 with their defining mutation, according to the ISOGG tree (as of January 2020). Note that the descent-based identifiers may be subject to change, as new SNPs are discovered that augment and further refine the tree. For brevity, only the first three levels of subclades are shown.
- J2 M172/Page28/PF4908, L228/PF4895/S321
- J2a M410, L152, L212/PF4988, L559/PF4986
- J2a1 DYS413≤18, L26/Page55/PF5110/S57, F4326/L27/PF5111/S396
- J2a1a M47, M322
- J2a1b M67/PF5137/S51
- J2a1c M68
- J2a1d M319
- J2a1e M339
- J2a1f M419
- J2a1g P81/PF4275
- J2a1h L24/S286, L207.1
- J2a1i L88.2, L198
- J2a2 L581/PF5026/S398
- J2a2a P279/PF5065
- J2a1 DYS413≤18, L26/Page55/PF5110/S57, F4326/L27/PF5111/S396
- J2b M12
- J2b1 M205
- J2b1a~ A11525, PH4306, Y22059, Y22060, Y22061, Y22062, Y22063
- J2b1b~ CTS1969
- J2b2~ CTS2622/Z1827, CTS11335/Z2440, Z575
- J2b2a M241
- J2b1 M205
- J2a M410, L152, L212/PF4988, L559/PF4986
See also
Genetics
Other Y-DNA J Subclades
Y-DNA Backbone Tree
References
- ↑ 1.0 1.1 "YFull YTree v7.05.00". https://yfull.com/tree/J2/.
- ↑ The extent of differentiation of Hg J, observed both with the biallelic and microsatellite markers, points to the Middle East as its likely homeland. In this area, J-M172 and J-M267 are equally represented and show the highest degree of internal variation, indicating that it is most likely that these subclades also arose in the Middle East. (Di Giacomo 2004)
- ↑ Shou, Wei-Hua; Qiao, En-Fa; Wei, Chuan-Yu; Dong, Yong-Li; Tan, Si-Jie; Shi, Hong; Tang, Wen-Ru; Xiao, Chun-Jie (2010). "Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians". Journal of Human Genetics 55 (5): 314–322. doi:10.1038/jhg.2010.30. PMID 20414255.
- ↑ "Afghanistan's ethnic groups share a Y-chromosomal heritage structured by historical events". PLOS ONE 7 (3): e34288. 2012. doi:10.1371/journal.pone.0034288. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- ↑ "Population Structure in the Mediterranean Basin: A Y Chromosome Perspective" (in en). Annals of Human Genetics 70 (2): 207–225. 2006. doi:10.1111/j.1529-8817.2005.00224.x. PMID 16626331.
- ↑ "Y haplogroup J in Iran by Alfred A. Aburto Jr.". http://archiver.rootsweb.ancestry.com/th/read/GENEALOGY-DNA/2006-06/1151592332.
- ↑ 7.0 7.1 7.2 "Трофимова Наталья Вадимовна, ИЗМЕНЧИВОСТЬ МИТОХОНДРИАЛЬНОЙ ДНК И Y-ХРОМОСОМЫВ ПОПУЛЯЦИЯХ ВОЛГО-УРАЛЬСКОГО РЕГИОНА, 03.02.07". http://ibg.anrb.ru/disovet/zashita/2014/09Trofimova/09TrofimovaDiser.pdf.
- ↑ A genetic study published led by Firasat (2007) on Kalash individuals found high and diverse frequencies.
- ↑ 9.0 9.1 Zalloua & Wells: National Geographic Magazine, October 2004. [1] and [2] .
- ↑ 10.0 10.1 "N. Al-Zahery et al. "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations" (2003)". Family Tree DNA. http://www.familytreedna.com/pdf/Al_Zahery.pdf.
- ↑ "Large-scale recent expansion of European patrilineages shown by population resequencing". Nature Communications 6: 7152. May 2015. doi:10.1038/ncomms8152. PMID 25988751. Bibcode: 2015NatCo...6.7152B.
- ↑ 12.0 12.1 "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area". American Journal of Human Genetics 74 (5): 1023–34. May 2004. doi:10.1086/386295. PMID 15069642.
- ↑ "[Homepage"]. http://www.yfull.com/.
- ↑ "Upper Palaeolithic genomes reveal deep roots of modern Eurasians". Nature Communications 6: 8912. November 2015. doi:10.1038/ncomms9912. PMID 26567969. Bibcode: 2015NatCo...6.8912J.
- ↑ Lazaridis, Iosif; Nadel, Dani; Rollefson, Gary; Merrett, Deborah C.; Rohland, Nadin; Mallick, Swapan; Fernandes, Daniel; Novak, Mario et al. (August 2016). "Genomic insights into the origin of farming in the ancient Near East". Nature 536 (7617): 419–424. doi:10.1038/nature19310. PMID 27459054. Bibcode: 2016Natur.536..419L.
- ↑ "The spread of the bull" (in en). 2018-07-15. https://aratta.wordpress.com/2018/07/15/the-spread-of-the-bull/.
- ↑ Al-Zahery, N. (2003). "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations.". Molecular Phylogenetics and Evolution 28 (3): 458–72. doi:10.1016/s1055-7903(03)00039-3. PMID 12927131.
- ↑ "The Y Chromosome Pool of Jews as Part of the Genetic Landscape of the Middle East". American Journal of Human Genetics 69 (5): 1095–1112. Nov 2001. doi:10.1086/324070. PMID 11573163. See especially Figure Six. Semino 2000 is a source which also states that Eu 9 descends from Eu 10 (Eu 10 is a different subclade of Haplogroup J (mtDNA)).
- ↑ "Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe". Human Genetics 115 (5): 357–71. 2004. doi:10.1007/s00439-004-1168-9. PMID 15322918.
- ↑ Al-Zahery, N.; Semino, O.; Benuzzi, G.; Magri, C.; Passarino, G.; Torroni, A.; Santachiara-Benerecetti, A. S. (2003-09-01). "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations". Molecular Phylogenetics and Evolution. Special Issue: Papers presented at the Mammalian Phylogeny symposium during the 2002 Annual Meeting of the Society for Molecular Biology and Evolution, Sorrento, Italy, June 13–16, 2002 28 (3): 458–472. doi:10.1016/S1055-7903(03)00039-3. ISSN 1055-7903. https://www.sciencedirect.com/science/article/pii/S1055790303000393.
- ↑ 21.0 21.1 Liu Shuhu; Nizam Yilihamu; Rabiyamu Bake; Abdukeram Bupatima; Dolkun Matyusup (2018). "Yìngyòng Y rǎnsètǐ SNP duì Xīnjiāng sān gè gélí rénqún yíchuán duōyàng xìng de yánjiū" (in zh). Rénlèixué xuébào / Acta Anthropologica Sinica 37 (1): 146–156. doi:10.16359/j.cnki.cn11-1963/q.2017.0067. http://www.anthropol.ac.cn/EN/Y2018/V37/I01/146.
- ↑ Shou, Wei-Hua; Qiao, Wn-Fa; Wei, Chuan-Yu; Dong, Yong-Li; Tan, Si-Jie; Shi, Hong; Tang, Wen-Ru; Xiao, Chun-Jie (2010). "Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians". J Hum Genet 55 (5): 314–322. doi:10.1038/jhg.2010.30. PMID 20414255.
- ↑ Allentoft (2015). "Population genomics of Bronze Age Eurasia". Nature 522 (7555): 167–172. doi:10.1038/nature14507. PMID 26062507. Bibcode: 2015Natur.522..167A. https://depot.ceon.pl/handle/123456789/13155. Retrieved 2020-01-21.
- ↑ C. Rоttensteiner, J2a2-PH3085, SK1403: Ancient Altai, modern Uygur and Turkish , J2-M172 Haplogroup Research.
- ↑ F. Immanuel, Codes for Gedmatch Results, Ancient DNA page , F999962 for RISE504, Kytmanovo sample, and F999965 for RISE602, Sary-bel sample.
- ↑ Al-Zahery et al, 2011, Additional file 3. Absolute frequencies of Y-chromosome haplogroups and sub-haplogroups in the 48 populations included in the PCA. Note: Only 37 of 154 samples (24%) are J2 in Iraq according to the list of Al-Zahery 2011. 43.6% is the frequency of J2 among all J haplogroup Iraqis, not all haplogroups.
- ↑ Abu-Amero (2009), Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions , Quote: The most abundant haplogroups in Saudi Arabia, J1-M267 (42%), J2-M172 (14%), E1-M2 (8%), R1-M17 (5%) and K2-M184 (5%) are also well represented in other Arabian populations (Table (Table1).1).
- ↑ 28.0 28.1 28.2 "Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup". Scientific Reports 6: 19157. January 2016. doi:10.1038/srep19157. PMID 26754573. Bibcode: 2016NatSR...619157S.
- ↑ Herrera, Rene J.; Garcia-Bertrand, Ralph (2018) (in en). Ancestral DNA, Human Origins, and Migrations. Academic Press. p. 250. ISBN 978-0-12-804128-4. https://books.google.com/books?id=ZF1gDwAAQBAJ&q=Ancestral+DNA+Human+Origins+and+Migrations+J2b-M102+South+Asia&pg=PA250.
- ↑ 30.0 30.1 30.2 30.3 "Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists". American Journal of Human Genetics 78 (2): 202–21. February 2006. doi:10.1086/499411. PMID 16400607.
- ↑ "Population differentiation of southern Indian male lineages correlates with agricultural expansions predating the caste system". PLOS ONE 7 (11): e50269. 2012. doi:10.1371/journal.pone.0050269. PMID 23209694. Bibcode: 2012PLoSO...750269A.
- ↑ "Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA". BMC Genetics 7: 42. August 2006. doi:10.1186/1471-2156-7-42. PMID 16893451.
- ↑ "The Indian origin of paternal haplogroup R1a1* substantiates the autochthonous origin of Brahmins and the caste system". Journal of Human Genetics 54 (1): 47–55. January 2009. doi:10.1038/jhg.2008.2. PMID 19158816.
- ↑ 34.0 34.1 "Y-Chromosomal DNA Variation in Pakistan". Am. J. Hum. Genet. 70 (5): 1107–24. May 2002. doi:10.1086/339929. PMID 11898125.
- ↑ "Indian Siddis: African Descendants with Indian Admixture". Am. J. Hum. Genet. 89 (1): 154–61. 2011. doi:10.1016/j.ajhg.2011.05.030. PMID 21741027.
- ↑ Toomas Kivisild; Siiri Rootsi; Mait Metspalu; Ene Metspalu; Juri Parik; Katrin Kaldma; Esien Usanga; Sarabjit Mastana et al. (2002). "The Genetics of Language and Farming Spread in India". Examining the farming/language dispersal hypothesis. McDonald Institute monographs. McDonald Institute for Archaeological Research. ISBN 9781902937205. http://evolutsioon.ut.ee/publications/Kivisild2003a.pdf.
- ↑ "Ancestry of Maldivian Islanders in Light of Population Genetics: Maldivian Ancestry in light of Genetics". May 24, 2013. http://maldives-ancestry.blogspot.com/2013/05/maldivian-ancestry-in-light-of-genetics.html.
- ↑ 38.0 38.1 38.2 Gyaneshwer Chaubey, Manvendra Singh, Federica Crivellaro, et al., "Unravelling the distinct strains of Tharu ancestry." European Journal of Human Genetics (2014), 1–9. doi:10.1038/ejhg.2014.36
- ↑ Simona Fornarino, Maria Pala, Vincenza Battaglia, et al., "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation." BMC Evolutionary Biology 2009, 9:154. doi:10.1186/1471-2148-9-154
- ↑ Singh, Sakshi; Singh, Ashish; Rajkumar, Raja; Sampath Kumar, Katakam; Kadarkarai Samy, Subburaj; Nizamuddin, Sheikh; Singh, Amita; Ahmed Sheikh, Shahnawaz et al. (2016-01-12). "Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup" (in en). Scientific Reports 6 (1): 19157. doi:10.1038/srep19157. ISSN 2045-2322.
- ↑ "Ossetian DNA Project - Y-DNA Classic Chart". familytreedna. https://www.familytreedna.com/public/Ossetian?iframe=yresults.
- ↑ "Haplogroup J2 (Y-DNA)". http://thegeneticatlas.com/J2_Y-DNA.htm.
- ↑ "Y-DNA Haplotree". https://www.familytreedna.com/y-dna-haplotree.aspx. Family Tree DNA uses the Y-Chromosome Consortium tree and posts it on their website.
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H. et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics 68 (2): 432–443. doi:10.1086/318205. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics 69 (3): 615–628. doi:10.1086/323299. PMID 11481588.
- Karafet, T. M.; Mendez, F. L.; Meilerman, M. B.; Underhill, P. A.; Zegura, S. L.; Hammer, M. F. (2008), "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree", Genome Research 18 (5): 830–8, doi:10.1101/gr.7172008, PMID 18385274
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.
Notes
- ^ Renfrew AC (1998). Archaeology and language: the puzzle of Indo-European origins (Pimlico ed.). London: Pimlico. ISBN 978-0-7126-6612-1.
- ^ "The Y chromosome pool of Jews as part of the genetic landscape of the Middle East". Am. J. Hum. Genet. 69 (5): 1095–112. 2001. doi:10.1086/324070. PMID 11573163.
- ^ "A multistep process for the dispersal of a Y chromosomal lineage in the Mediterranean area". Ann. Hum. Genet. 65 (Pt 4): 339–49. 2001. doi:10.1046/j.1469-1809.2001.6540339.x. PMID 11592923. https://art.torvergata.it/bitstream/2108/44448/1/Malaspina_AHG_2001.pdf.
Bibliography
- "Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions". BMC Genetics 10: 59. 2009. doi:10.1186/1471-2156-10-59. PMID 19772609.
- Adams, SM; Bosch, E; Balaresque, PL; Ballereau, SJ; Lee, AC; Arroyo, E; López-Parra, AM; Aler, M et al. (2008). "The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula". American Journal of Human Genetics 83 (6): 725–36. doi:10.1016/j.ajhg.2008.11.007. PMID 19061982.
- "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations". Molecular Phylogenetics and Evolution 28 (3): 458–72. 2003. doi:10.1016/S1055-7903(03)00039-3. PMID 12927131.
- "In search of the genetic footprints of Sumerians: A survey of Y-chromosome and mtDNA variation in the Marsh Arabs of Iraq". BMC Evolutionary Biology 11: 288. 2011. doi:10.1186/1471-2148-11-288. PMID 21970613.
- "A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa". The American Journal of Human Genetics 75 (2): 338–45. 2004. doi:10.1086/423147. PMID 15202071.
- Ashirbekov, E. E. (2017). "Distribution of Y-Chromosome Haplogroups of the Kazakh from the South Kazakhstan, Zhambyl, and Almaty Regions". Reports of the National Academy of Sciences of the Republic of Kazakhstan 6 (316): 85–95. http://nblib.library.kz/elib/library.kz/jurnal/%D0%94%D0%BE%D0%BA%D0%BB%D0%B0%D0%B4_06_2017%20(2)/12-%20Biology%20E.E.Ashirbekov0617.pdf. Retrieved 2023-05-26.
- "Parallel Evolution of Genes and Languages in the Caucasus Region". Molecular Biology and Evolution 28 (10): 2905–20. 2011. doi:10.1093/molbev/msr126. PMID 21571925.
- "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe". European Journal of Human Genetics 17 (6): 820–30. 2008. doi:10.1038/ejhg.2008.249. PMID 19107149.
- Behar, Doron M.; Garrigan, Daniel; Kaplan, Matthew E.; Mobasher, Zahra; Rosengarten, Dror; Karafet, Tatiana M.; Quintana-Murci, Lluis; Ostrer, Harry et al. (1 March 2004). "Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations". Human Genetics 114 (4): 354–365. doi:10.1007/s00439-003-1073-7. PMID 14740294.
- "Y-chromosome diversity characterizes the Gulf of Oman". European Journal of Human Genetics 16 (3): 374–86. 2007. doi:10.1038/sj.ejhg.5201934. PMID 17928816.
- "Population Structure in the Mediterranean Basin: A Y Chromosome Perspective". Annals of Human Genetics 70 (2): 207–25. 2006. doi:10.1111/j.1529-8817.2005.00224.x. PMID 16626331.
- "Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic–Neolithic encounter". Molecular Phylogenetics and Evolution 44 (1): 228–39. 2007. doi:10.1016/j.ympev.2006.11.030. PMID 17275346.
- "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics 114 (2): 127–48. 2004. doi:10.1007/s00439-003-1031-4. PMID 14586639.
- "Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects". Molecular Phylogenetics and Evolution 28 (3): 387–95. 2003. doi:10.1016/S1055-7903(03)00016-2. PMID 12927125.
- "Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations". European Journal of Human Genetics 18 (3): 354–63. 2009. doi:10.1038/ejhg.2009.168. PMID 19809480.
- "Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast". Annals of Human Genetics 73 (6): 568–81. 2009. doi:10.1111/j.1469-1809.2009.00538.x. PMID 19686289. This paper reported results from several studies : Di Giacomo 2003, Al-Zahery 2003, Flores 2004, Cinnioglu 2004, Capelli 2005, Goncalves 2005, Zalloua 2008, Cadenas 2008
- Fadhlaoui-Zid, Karima (2014). "Sousse: extreme genetic heterogeneity in North Africa". Journal of Human Genetics 60 (1): 41–49. doi:10.1038/jhg.2014.99. PMID 25471516.
- Fregel, Rosa; Gomes, Verónica; Gusmão, Leonor; González, Ana M; Cabrera, Vicente M; Amorim, António; Larruga, Jose M (2009). "Demographic history of Canary Islands male gene-pool: replacement of native lineages by European". BMC Evolutionary Biology 9 (1): 181. doi:10.1186/1471-2148-9-181. PMID 19650893.
- Firasat, Sadaf; Khaliq, Shagufta; Mohyuddin, Aisha; Papaioannou, Myrto; Tyler-Smith, Chris; Underhill, Peter A.; Ayub, Qasim (2007). "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan". European Journal of Human Genetics 15 (1): 121–126. doi:10.1038/sj.ejhg.5201726. PMID 17047675.
- "Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe". Human Genetics 115 (5): 357–71. 2004. doi:10.1007/s00439-004-1168-9. PMID 15322918.
- Kivisild, Toomas, ed (2012). "Ancient Migratory Events in the Middle East: New Clues from the Y-Chromosome Variation of Modern Iranians". PLOS ONE 7 (7): e41252. doi:10.1371/journal.pone.0041252. PMID 22815981. Bibcode: 2012PLoSO...741252G.
- Guo, Yuxin; Xia, Zhiyu; Cui, Wei; Chen, Chong; Jin, Xiaoye; Zhu, Bofeng (2020-05-18). "Joint Genetic Analyses of Mitochondrial and Y-Chromosome Molecular Markers for a Population from Northwest China". Genes 11 (5): 564. doi:10.3390/genes11050564. PMID 32443545.
- Kayser, Manfred, ed (2011). "Afghanistan's Ethnic Groups Share a Y-Chromosomal Heritage Structured by Historical Events". PLOS ONE 7 (3): e34288. doi:10.1371/journal.pone.0034288. PMID 22470552. Bibcode: 2012PLoSO...734288H.
- "Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages". Antiquity 76 (293): 707–714. 2002. doi:10.1017/s0003598x00091158.
- "Differential Y-chromosome Anatolian Influences on the Greek and Cretan Neolithic". Annals of Human Genetics 72 (2): 205–14. 2008. doi:10.1111/j.1469-1809.2007.00414.x. PMID 18269686.
- Krahn; FTDNA (2003). "Genomic Research Center Draft Tree (AKA Y-TRee)". http://ytree.ftdna.com/index.php?name=Draft&parent=root.
- "The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations". The American Journal of Human Genetics 74 (3): 532–44. 2004. doi:10.1086/382286. PMID 14973781.
- "A multistep process for the dispersal of a Y chromosomal lineage in the Mediterranean area". Annals of Human Genetics 65 (4): 339–49. 2001. doi:10.1046/j.1469-1809.2001.6540339.x. PMID 11592923.
- "Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau". European Journal of Human Genetics 15 (4): 485–93. 2007. doi:10.1038/sj.ejhg.5201769. PMID 17264870.
- "Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome". Human Genetics 112 (3): 255–61. 2003. doi:10.1007/s00439-002-0874-4. PMID 12596050.
- "Mitochondrial DNA and Y-Chromosome Variation in the Caucasus". Annals of Human Genetics 68 (3): 205–21. 2004. doi:10.1046/j.1529-8817.2004.00092.x. PMID 15180701.
- "The Y Chromosome Pool of Jews as Part of the Genetic Landscape of the Middle East". The American Journal of Human Genetics 69 (5): 1095–112. 2001. doi:10.1086/324070. PMID 11573163.
- Nogueiro, I.; Manco, L.; Gomes, V.; Amorim, A.; Gusmão, L. (2010). "Phylogeographic analysis of paternal lineages in NE Portuguese Jewish communities". American Journal of Physical Anthropology 141 (3): 373–381. doi:10.1002/ajpa.21154. PMID 19918998.
- "Iran: Tricontinental Nexus for Y-Chromosome Driven Migration". Human Heredity 61 (3): 132–43. 2006. doi:10.1159/000093774. PMID 16770078.
- Robino, C.; Crobu, F.; Di Gaetano, C.; Bekada, A.; Benhamamouch, S.; Cerutti, N.; Piazza, A.; Inturri, S. et al. (2008). "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample". International Journal of Legal Medicine 122 (3): 251–255. doi:10.1007/s00414-007-0203-5. PMID 17909833.
- "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males". European Journal of Human Genetics 13 (7): 856–66. 2005. doi:10.1038/sj.ejhg.5201390. PMID 15756297.
- "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective". Science 290 (5494): 1155–9. 2000. doi:10.1126/science.290.5494.1155. PMID 11073453. Bibcode: 2000Sci...290.1155S.
- "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area". The American Journal of Human Genetics 74 (5): 1023–34. 2004. doi:10.1086/386295. PMID 15069642.
- "Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists". The American Journal of Human Genetics 78 (2): 202–21. 2006. doi:10.1086/499411. PMID 16400607.
- "Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-Chromosome and mitochondrial DNA sequence Variation". Human Mutation 24 (3): 248–60. 2004. doi:10.1002/humu.20077. PMID 15300852.
- Soodyall, H (2013). "Lemba origins revisited: Tracing the ancestry of Y chromosomes in South African and Zimbabwean Lemba". South African Medical Journal 103 (12): 1009–1013. doi:10.7196/SAMJ.7297. PMID 24300649. http://www.ajol.info/index.php/samj/article/view/98362. Retrieved 9 May 2014.
- "J1-M267 Y lineage marks climate-driven pre-historical human displacements". European Journal of Human Genetics 17 (11): 1520–4. 2009. doi:10.1038/ejhg.2009.58. PMID 19367321.
- "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–61. 2000. doi:10.1038/81685. PMID 11062480.
- "The Eurasian Heartland: A continental perspective on Y-chromosome diversity". Proceedings of the National Academy of Sciences 98 (18): 10244–9. 2001. doi:10.1073/pnas.171305098. PMID 11526236. Bibcode: 2001PNAS...9810244W.
- "The Caucasus as an asymmetric semipermeable barrier to ancient human migrations". Molecular Biology and Evolution 29 (1): 359–65. 2012. doi:10.1093/molbev/msr221. PMID 21917723.
- "Y-Chromosomal Diversity in Lebanon is Structured by Recent Historical Events". The American Journal of Human Genetics 82 (4): 873–82. 2008l. doi:10.1016/j.ajhg.2008.01.020. PMID 18374297.
- "Afghan Hindu Kush: where Eurasian sub-continent gene flows converge". PLOS ONE 8 (10): e76748. 2013. doi:10.1371/journal.pone.0076748. PMID 24204668. Bibcode: 2013PLoSO...876748D.
- "Y-chromosome distributions among populations in Northwest China identify significant contribution from Central Asian pastoralists and lesser influence of western Eurasians". Journal of Human Genetics 55 (5): 314–22. 2010. doi:10.1038/jhg.2010.30. PMID 20414255.
External links
- Migration of Indians Across Continents spanning generations: A Case History of the Saluja Family.
- In Lebanon DNA may yet heal rifts
Phylogenetic notes
- ↑ This table shows the historic names for J-M172 in published peer reviewed literature. Note that in Semino 2000 Eu09 is a subclade of Eu10 and in Karafet 2001 24 is a subclade of 23.
YCC 2002/2008 (Shorthand) J-M172 Jobling and Tyler-Smith 2000 9 Underhill 2000 VI Hammer 2001 Med Karafet 2001 24 Semino 2000 Eu9 Su 1999 H4 Capelli 2001 B YCC 2002 (Longhand) J2* YCC 2005 (Longhand) J2 YCC 2008 (Longhand) J2 YCC 2010r (Longhand) J2 - ↑ This table shows the historic names for J-P209 (AKA J-12f2.1 or J-M304) in published peer reviewed literature. Note that in Semino 2000 Eu09 is a subclade of Eu10 and in Karafet 2001 24 is a subclade of 23.
YCC 2002/2008 (Shorthand) J-P209
(AKA J-12f2.1 or J-M304)Jobling and Tyler-Smith 2000 9 Underhill 2000 VI Hammer 2001 Med Karafet 2001 23 Semino 2000 Eu10 Su 1999 H4 Capelli 2001 B YCC 2002 (Longhand) J* YCC 2005 (Longhand) J YCC 2008 (Longhand) J YCC 2010r (Longhand) J - ↑ "ISOGG 2018 Y-DNA Haplogroup J". http://www.isogg.org/tree/ISOGG_HapgrpJ.html.
 |

