Biology:Hya RNA motif
From HandWiki
| hya | |
|---|---|
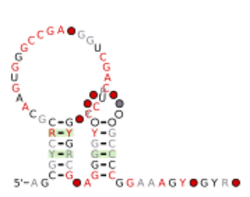 Consensus secondary structure and sequence conservation of hya RNA | |
| Identifiers | |
| Symbol | hya |
| Rfam | RF02992 |
| Other data | |
| RNA type | Cis-reg |
| SO | 0005836 |
| PDB structures | PDBe |
The hya RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] hya motif RNAs are found in Actinomycetota.
hya motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. Indeed, the RNAs are upstream of multiple genes that encode non-homologous proteins. If all examples of the RNA were upstream of homologous genes, there is the possibility that the RNAs were conserved in that position simply by inheritance. The non-homology of the genes downstream of hya RNAs makes this scenario less likely. The genes presumably regulated by hya RNAs are subunits of nickel-iron hydrogenase I.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |

