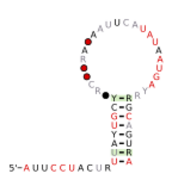Biology:MalK RNA motifs
malK RNA motifs are conserved RNA structures that were discovered by bioinformatics.[1] They are defined by being consistently located upstream of malK genes, which encode an ATPase that is used by transporters whose ligand is likely a kind of sugar. Most of these genes are annotated either as transporting maltose or glycerol-3-phosphate, however the substrate of the transporters associated with malK motif RNAs has not been experimentally determined. All known types of malK RNA motif are generally located nearby to the Shine-Dalgarno sequence of the downstream gene.
Given their positions upstream of genes and proximal to Shine-Dalgarno sequences, malK RNA motifs likely function as cis-regulatory elements, to regulate their associated malK genes. It is possible that they bind a sugar directly, and therefore function as riboswitches, but it is equally possible that regulation is mediated via a protein factor, or some other mechanism.
Three class of malK RNA motif have been found, and each class exhibits a completely unrelated structure, in terms of different conserved sequence features and a different conserved secondary structure. malK-I motifs are found in Clostridia, while malK-II motifs are found in organisms within the genus Vibrio. malK-III motifs have not yet (as of 2018) been identified in any classified organism but are known only from sequences isolated from metagenomic environmental samples.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |