Biology:Membrane bound polyribosome
In cell biology, membrane bound polyribosomes are attached to a cell's endoplasmic reticulum.[1] When certain proteins are synthesized by a ribosome they can become "membrane-bound". The newly produced polypeptide chains are inserted directly into the endoplasmic reticulum by the ribosome and are then transported to their destinations. Bound ribosomes usually produce proteins that are used within the cell membrane or are expelled from the cell via exocytosis.[2]
Background
A membrane-bound polyribosome, as the name suggests, is composed of multiple ribosomes that are associated with a membrane. Proteins are synthesized via messenger ribonucleic acid (mRNA) from the nucleus being released either into the cytoplasm or into the rough endoplasmic reticulum.[3] The rough endoplasmic reticulum branches off of the cell nucleus, has multiple cisternae or layered folds that have interstitial space for protein extrusion.[3] Ribosomes are located in both the cytosol, cellular fluid, or rough endoplasmic reticulum and attach to this ribonucleic acid by separation and re-association of subunits around the messenger ribonucleic acid.[3] In eukaryotic cells, the small subunit (40S) stays on one side and translates the messenger ribonucleic acid while the large subunit (60S) goes codon by codon down the mRNA and attaches each amino acids coded for making a polypeptide.[3][4] A polysome is when multiple ribosomes attach to the same strand of messenger ribonucleic acid.[3] The polypeptides ribosomes produce go on to be cell structural proteins, enzymes, and many other things.[3] Ribosomes can also sometimes be associated with chloroplasts and mitochondria but these are not membrane bound.[3]
Origin
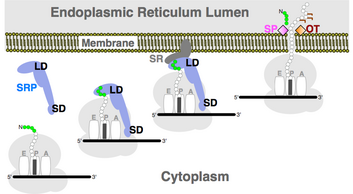
Free-floating ribosomes can become membrane bound through a process called translocation.[5] Through translocation, ribosomes that are found in the cytosol producing proteins are moved and attached to the membrane.[3] This process is responsible for development of the rough endoplasmic reticulum.[3] First, ribosomes begin protein synthesis at the N-terminus.[3] The first of the polypeptide may actually be a signal sequence that tells the ribosome that the protein must be extruded into the rough endoplasmic reticulum.[3] The signal sequence triggers translocation by binding with a signal recognition particle (SRP) also located in the cytosol.[3] The signal recognition particle allows recognition and binding via a signal recognition particle receptor on the target membrane’s surface.[3] The signal recognition particle receptor and signal recognition particle are both attached to a translocon and bound with guanosine triphosphate (GTP).[3] This guanosine triphosphate is phosphorylated for energy and opens the translocon allowing the ribosome to attach via its 60S subunit and its signal sequence to enter the lumen, or institial space of the rough endoplasmic reticulum.[3] The signal recognition particle and signal recognition particle receptor detach and can be recycled.[3] The signal sequence is cleaved inside the lumen of the rough endoplasmic reticulum and the ribosome continues to produce the protein into the endoplasmic reticulum where it is folded.[3] Upon protein synthesis completion, the translocon closes and the ribosome detaches. It is important to remember that during translocation, translation briefly stops until binding with the membrane is finished.[3] It is also important to remember that ribosomes can associate and dissociate with the endoplasmic reticulum as need for protein synthesis.[6] After synthesis into the rough endoplasmic reticulum, proteins may travel to the end of the rough endoplasmic reticulum where they are exocytosed, or packaged into small vesicles formed via cleavage of the membrane of the rough endoplasmic reticulum. These vesicles are sent to the Golgi apparatus for sorting and release as needed by the cell.[3] Some proteins are made to be released immediately as the cell is in constant need of them while some proteins are store for immediate release upon signal.[3]
The idea that translation and translocation occur simultaneously except in some yeasts was confirmed via microsomes.[3] Microsomes are small vesicles of rough endoplasmic reticulum's membranes formed after disruption of the organelle via homogenation.[7] Homogenation is physical disruption of cells.[7] Microsomes form after homogenization because of the membrane nature of the endoplasmic reticulum.[3] In a lipid bilayer, hydrophobic tails must come together and be hydrophilic head must face the external aqueous environment.[3] In an experiment, proteins were synthesized via ribosomes with microsomes added simultaneously and with microsomes added after synthesis.[3] In the group where microsomes were added simultaneously, the proteins were synthesized into the microsome with the signal sequence cleaved.[3] In the group where microsomes were added post protein synthesis, the proteins were located outside the microsome and retained their signal sequence.[3] Therefore, it is possible to tell if a protein has been extruded into a microsome by its length (lack of a N-terminal signal protein if extruded), resistance to proteases, lack of resistance to proteases in the presence of detergents, and glycosylation.[3] It was confirmed that non-extruded proteins are longer via SDS-page of proteins in the presence of and without microsomes.[3] Protease resistance is due to the characteristics of the surrounding endoplasmic reticulum.[3] And glycosylation occurs via glycosyltransferases to help with folding and stabilization of proteins.[3]
Significance
The cleavage of a signal protein, resistance to proteases, and glycosylation provided by the endoplasmic reticulum to membrane-bound polyribosomes allows for more effective protein production.[3] Presence of the signal protein makes the protein bulkier, a different shape, and harder to store until the unusable signal sequence can be cleaved.[3] The protection from proteases due to protection by the endoplasmic reticulum prevents the protein from being degraded as it is formed.[3] Extrusion in the endoplasmic reticulum also makes sure that the protein folds correctly.[3] Resident endoplasmic reticulum proteins like binding protein (BiP), protein disulfide isomerase (PDI), and glycosyltransferases (GTs) are all responsible for ensuring correct protein folding and stabilization as the protein is assembled.[3] Binding protein can actively help fold or prevent folding of proteins while protein disulfide isomerase promotes the formation of disulfide bridges.[3] Glycosyltransferases promotes the glycosylation or incorporation of a carbohydrate to improve rigidity or structure of a protein.[3] Failure of proteins to fold correctly may result in the unfolded protein response.[3] Unfolded proteins cause swelling of the endoplasmic reticulum as more unfolded proteins continue to be produced.[3] The unfolded protein response can result in endoplasmic reticulum stress, phosphorylation of PERK, phosphorylation of elF2a, down regulation of protein production, and possibly apoptosis.[3] Apoptosis of affected cells may result in a disease like Amylotrophic Lateral Sclerosis (ALS).[3] In Amylotrophic Lateral Sclerosis, dendritic cells undergo endoplasmic reticulum stress because of misfiled SOD1 proteins and apoptose resulting in lack of nerve transmission and loss of muscle control.[3] Eventually, those with Amylotrophic Lateral Sclerosis die because of lack of nerve impulses to signal breathing or heart ventricle contraction.
References
- ↑ Seiser, Robert M.; Nicchitta, Christopher V. (2000). "The Fate of Membrane-bound Ribosomes Following the Termination of Protein Synthesis". Journal of Biological Chemistry 275 (43): 33820–33827. doi:10.1074/jbc.M004462200. PMID 10931837. http://www.jbc.org/content/275/43/33820.full. Retrieved 29 October 2017.
- ↑ Bhardwaj, Uma; Bhardwaj, Ravindra. Biochemistry for Nurses. Pearson Education India. p. 11. ISBN 9788131795286. https://books.google.com/books?id=93yeKr9W9TwC.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 3.24 3.25 3.26 3.27 3.28 3.29 3.30 3.31 3.32 3.33 3.34 3.35 3.36 3.37 3.38 3.39 3.40 3.41 Lodish, Harvey (April 1, 2016) (in English). Molecular Cell Biology (8th ed.). N/A: W. H. Freeman. pp. Whole book. ISBN 978-1464183393.
- ↑ Gregory, Brian; Rahman, Nusrat; Bommakanti, Ananth; Shamsuzzaman, Md; Thapa, Mamata; Lescure, Alana; Zengel, Janice M; Lindahl, Lasse (2019-03-05). "The small and large ribosomal subunits depend on each other for stability and accumulation". Life Science Alliance 2 (2): e201800150. doi:10.26508/lsa.201800150. ISSN 2575-1077. PMID 30837296.
- ↑ Fewell, Sheara W.; Brodsky, Jeffrey L. (2013) (in en). Entry into the Endoplasmic Reticulum: Protein Translocation, Folding and Quality Control. Landes Bioscience. https://www.ncbi.nlm.nih.gov/books/NBK6210/.
- ↑ Sanvictores, Terrence; Davis, Donald D. (2022), "Histology, Rough Endoplasmic Reticulum", StatPearls (Treasure Island (FL): StatPearls Publishing), PMID 33085273, http://www.ncbi.nlm.nih.gov/books/NBK563126/, retrieved 2022-12-07
- ↑ 7.0 7.1 de Araújo, Mariana E. G.; Lamberti, Giorgia; Huber, Lukas A. (2015-11-02). "Homogenization of Mammalian Cells". Cold Spring Harbor Protocols 2015 (11): 1009–1012. doi:10.1101/pdb.prot083436. ISSN 1559-6095. PMID 26527761. https://pubmed.ncbi.nlm.nih.gov/26527761/.
 |