Biology:PANO1
 Generic protein structure example |
PANO1 is a protein which in humans is encoded by the PANO1 gene. PANO1 is an apoptosis inducing protein that is able to regulate the function of tumor suppressor.[1] More specifically, P14ARF is a protein in which in humans is modulated by the PANO1 gene. P14ARF is known to function as a tumor suppressor.[2] When PANO1 is highly expressed in the cells, it is able to modulate p14ARF by stabilizing it and protecting it from degradation.[2] With a confidence level of 5 out of 5, PANO1 has been theorized to be expressed in the nucleolus of the cell.[3] PANO1 is an intron-less gene.[1] Intron-less genes only make up about 3% of the human genome.[4] A functional analysis of these types of genes revealed that they often have tissue-specific expression in tissues such as the nervous system and testis.[4] This kind of expression is commonly associated with neuropathies, disease, and cancer.[4] The tissue types that PANO1 has the highest expression in, are the cerebellum regions of the brain as well as pituitary and testis tissues.[5]
Gene
PANO1 is also known as Proapoptotic Nucleolar Protein 1, PANO, and Pre-mRNA-splicing Factor CW22-like. PANO1 is located on human chromosome 11 at positions 797,511-799,190 and is positioned on the + strand.[6] Its protein contains 1 exon and 215 amino acids.[1]
Transcripts and proteins
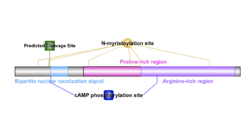
PANO1 has one isoform, isoform 1, located in the PONAB and PANTR species.[1] These isoforms have proteins with 215 and 216 amino acids, respectively.[1] No isoforms for the human PANO1 protein could be identified. Human PANO1 protein has a molecular weight of 22.8 kb and a theoretical, isoelectric point of 12.21.[1] From an analysis of the PANO1 protein, it was observed that the protein contains a low amount of lysine and a very low amount of asparagine when compared to other human proteins.[7] The same analysis indicated that the protein does not contain any hydrophobic or transmembrane regions.[7] PANO1 contains 2 cAMP phosphorylation sites, 6 N-myristoylation sites, 4 protein kinase C phosphorylation sites, 3 bipartite nuclear localization signals as well as arginine-rich and proline-rich regions. Using PSORTII, 3 discrimination of nuclear localization signals were identified.[8] Pat4 (RRRR) at position 200, Pat7 (RRRR) at position 201 and a bipartite (RKGTPTARCLGQRTKEK) at position 35.[8] Nuclear localization signals allow proteins to be able to enter the nucleus, but many nuclear proteins possess their own.[8] PANO1 overlaps solute carrier family 25 member 22 (SLC25A22).[3] A causal link between this solute carrier, when upregulated, has been strongly associated with an osteosarcoma's ability to proliferate.[9] In addition to promoting proliferation, SLC25A22 when expressed in vitro, progressed the cell cycle and inhibited apoptosis.[9] It is curious that PANO1, an apoptosis inducer, and SLC25A22, an apoptosis inhibiter, overlap one another.
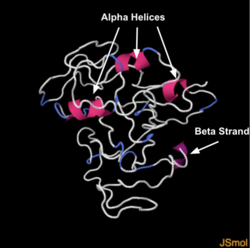
Structure
The structure of PANO1 is 82% disordered meaning the protein is able to move around easily.[10] The secondary structure reveals a beta strand at positions 3-9 as well as alpha helices incorporated throughout.[10]
Gene level regulation
Promoter
One promoter region was identified using Genomatix. It is located on the positive strand and is 1040 bp in length.[11] Its start site is located at 795633 and it ends at 796672.[11] It overlaps with the primary transcript which starts at 796633.[11] 440 transcription factors were identified within the promoter region.[11] To highlight some of relevance and importance to PANO1: Wilms tumor suppressor, spermatogenic zip 1 transcription factor, signal transducer and activator of transcription, pleomorphic adenoma gene, general transcription factor IIIA, stimulating protein 1, CCAAT/enhancer binding protein, GC box elements and HMG box-containing protein 1.
Expression
Like previously mentioned, PANO1 is detected to be most expressed in the cerebellum, pituitary and testis tissues.[5] Furthermore, NCBI Geo profiles indicated that PANO1 is highly expressed in B-lymphocytes, liposarcomas, and the testis cell lines.[12] It also appears that PANO1 is highly expressed in the androgen-sensitive LAPC-4 cell line with a rank of 97. PANO1 is biased to being expressed in androgen sensitive cells compared to androgen insensitive cells.[12]
Transcript level regulation
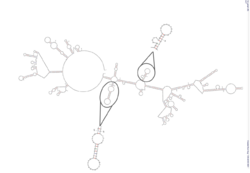
A predicted 3' UTR structure was generated using Unafold and depicts predicted stem loop structures.[13] Two stem loop structures are zoomed in on.
Protein level regulation
A possible cleavage site was identified between amino acids 33 and 34 as depicted in the PANO1 protein model.[1] As mentioned previously, 6 N-myristoylation sites, 2 cAMP phosphorylation sites, and 4 protein kinase C phosphorylation sites are also present.[1]
| Motif | Location (aa) |
|---|---|
| N-myristoylation site | 23-28 |
| N-myristoylation site | 33-38 |
| N-myristoylation site | 71-76 |
| N-myristoylation site | 111-116 |
| N-myristoylation site | 120-125 |
| N-myristoylation site | 186-191 |
| cAMP phosphorylation site | 35-38 |
| cAMP phosphorylation site | 136-139 |
| protein kinase C phosphorylation site | 34-36 |
| protein kinase C phosphorylation site | 40-42 |
| protein kinase C phosphorylation site | 57-59 |
| protein kinase C phosphorylation site | 191-193 |
Homology and evolutionary history
Orthologs and phylogenic tree
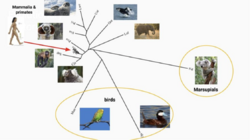
PANO1 orthologs were only able to be traced back in divergence to birds. Much more closely related orthologs include primates, as well as marsupial and placental mammals.[14] Specific examples of orthologs can be seen in the table below.
| Genus & species | Common name | Tax. group | Date of divergence (MYA) | Accession # | Seq. length (aa) | Seq. identity to HSA prot. (%) | Seq. identity to HSA prot. (#/100) | Seq. similarity to HSA prot. (%) | n | m |
| Homo sapiens | Human | human | 0 | XP_034607700.1 | 215 | 100 | 1 | 100 | 0 | 0 |
| Pongo abelii | Sumatran orangutan | primate | 15.76 | XP_024111240.1 | 216 | 96.3 | 0.963 | 100 | 0.037 | 3.770186718 |
| Sapajus apella | tufted capuchin | primate | 43.2 | XP_024111240.1 | 216 | 96.3 | 0.963 | 100 | 0.037 | 3.770186718 |
| Hylobates moloch | silvery gibbon | primate | 19.8 | XP_032005218.1 | 175 | 95.43 | 0.9543 | 81 | 0.0457 | 4.677719156 |
| Rhinopithecus roxellana | golden monkey | primate | 28.21 | XP_030773457.1 | 239 | 86.63 | 0.8663 | 86 | 0.1337 | 14.35240101 |
| Callithrix jacchus | white-tufted-eared marmoset | primate | 42.9 | XP_009007032.2 | 268 | 72.5 | 0.725 | 91 | 0.275 | 32.15836241 |
| Propithecus coquereli | Coquerel's sifaka | primate | 74.1 | XP_012507388.1 | 183 | 69.28 | 0.6928 | 71 | 0.3072 | 36.70139217 |
| Phascolarctos cinereus | koala | marsupial | 160 | XP_020836186.1 | 205 | 28.5 | 0.285 | 0.715 | 125.5266099 | |
| Leptonychotes weddellii | Weddell seal | placental | 96 | XP_030893169.1 | 182 | 47.88 | 0.4788 | 70 | 0.5212 | 73.64723053 |
| Phoca vitulina | harbor seal | placental | 96 | XP_032270736.1 | 264 | 54.55 | 0.5455 | 67 | 0.4545 | 60.60524737 |
| Loxodonta africana | African bush elephant | placental | 105 | XP_010600675.1 | 204 | 39 | 0.39 | 85 | 0.61 | 94.16085399 |
| Orcinus orca | killer whale | placental | 96 | XP_012391505.1 | 237 | 41.21 | 0.4121 | 91 | 0.5879 | 88.64892406 |
| Felis catus | house cat | placental | 96 | XP_023096045.1 | 217 | 59.86 | 0.5986 | 0.4014 | 51.31616836 | |
| Ursus arctos horribilis | grizzly bear | placental | 96 | XP_026346196.1 | 290 | 60.34 | 0.6034 | 53 | 0.3966 | 50.51749523 |
| Oxyura jamaicensis | ruddy duck | bird | 312 | XP_035173110.1 | 278 | 29.26 | 0.2926 | 0.7074 | 122.894879 | |
| Melopsittacus undulatus | budgerigar | bird | 312 | XP_033919672.1 | 228 | 28.73 | 0.2873 | 0.7127 | 124.7228313 |
Divergence
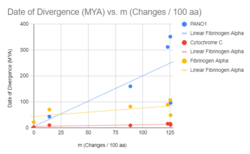
PANO1 was compared to two other genes, fibrinogen alpha chain as well as cytochrome C. The date of divergence as well as amino acid changes were tracked over many different species types to generate a divergence date vs. number of amino acids changes as seen to the right. PANO1 appears to diverge much more quickly than fibrinogen alpha and much more quickly than cytochrome c.
Interacting proteins
p14ARF is a protein that is a known tumor suppressor.[2] It does this by controlling cell proliferation and cell survival, however the mechanism for how this process is controlled/modulated remained unclear.[2] PANO1 has been identified to modulate and stabilize p14ARF by stabilizing it and protecting it from degradation in HeLa cells.[2] When PANO1 is over-expressed, as a direct result, p14ARF expression also increases.[2]
Clinical significance
When there is an abnormal expression of PANO1 in HeLa cells, scientists have seen a decrease in tumorigenicity in nude mice.[2] Additionally, other diseases that have been associated with PANO1 include hemochromatosis 2.[3]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 "PANO1 - Proapoptotic nucleolar protein 1 - Homo sapiens (Human) - PANO1 gene & protein" (in en). https://www.uniprot.org/uniprot/I0J062#family_and_domains.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 "A novel proapoptotic gene PANO encodes a post-translational modulator of the tumor suppressor p14ARF". Exp Cell Res 318 (3): 187–95. February 2012. doi:10.1016/j.yexcr.2011.10.019. PMID 22094112.
- ↑ 3.0 3.1 3.2 "PANO1". https://www.genecards.org/cgi-bin/carddisp.pl?gene=PANO1&keywords=PANO1.
- ↑ 4.0 4.1 4.2 "Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing". Biochem Biophys Res Commun 424 (1): 1–6. July 2012. doi:10.1016/j.bbrc.2012.06.092. PMID 22732409.
- ↑ 5.0 5.1 "Human BLAT Search". https://genome.ucsc.edu/cgi-bin/hgBlat.
- ↑ "GeneLoc Integrated Map for Chromosome 11: Search Results". https://genecards.weizmann.ac.il/geneloc-bin/display_map.pl?range_type=within_id&chr_nr=11&around_id=572477&around_marker=PANO1&start_bp=797511&end_bp=799190&plus_minus_Mb=4&match=exact&range=on&genes=on&enhancers=on&entity=gene#572477.
- ↑ 7.0 7.1 "SAPS < Sequence Statistics < EMBL-EBI". https://www.ebi.ac.uk/Tools/seqstats/saps/.
- ↑ 8.0 8.1 8.2 "PSORT II Prediction". https://psort.hgc.jp/form2.html.
- ↑ 9.0 9.1 "SLC25A22 Promotes Proliferation and Metastasis of Osteosarcoma Cells via the PTEN Signaling Pathway". Technol Cancer Res Treat 17: 1533033818811143. January 2018. doi:10.1177/1533033818811143. PMID 30482097.
- ↑ 10.0 10.1 "Phyre 2 Results for PANO1_phyre2". http://www.sbg.bio.ic.ac.uk/phyre2/phyre2_output/2f8624972f0d536e/summary.html.
- ↑ 11.0 11.1 11.2 11.3 "Genomatix Software Suite". https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=c8f0c29ba9b1c355fb5ff6042d142dd6;SHOW_ANNOTATION=PANO1;ELDORADO_VERSION=E36R2011.
- ↑ 12.0 12.1 "PANO1 - GEO Profiles - NCBI". https://www.ncbi.nlm.nih.gov/geoprofiles/?term=PANO1.
- ↑ "DNA folding results on 21Aug01-20-42-28 for 73.242.41.233". http://www.unafold.org/results/20/21Aug01-20-42-28/.
- ↑ "Protein BLAST: search protein databases using a protein query". https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
 |
