Biology:PRR32
 Generic protein structure example |
PRR32 is a protein that in humans is encoded by the CXorf64 (Chromosome X open reading frame 64) gene. It was also found that the homologs of the PRR32 gene is conserved in chimpanzee, Rhesus monkey, dog, cow, mouse, and rat. It was also found through ncbi that 82 organisms have orthologs with human gene PRR323.[1]
PRR32 (CXorf64) seems to be involved with a group of genes over-expressed in ALS (Amyotrophic lateral sclerosis), evident from a study aiming to study gene expression patterns in muscles from patients with amyotrophic lateral sclerosis and multifocal motor neuropathy.[2]
Gene
The gene is located on Chromosome X, at position Xq25, and is 2023 bases long (significantly small when compared to other genes). Its location is at the antisense (+) strand. As shown in the graphic below, it is flanked by upstream by the DKAF12l1 shown in green on the left and downstream by LOC10. CXorf64 is the blue column shown in the middle.[3]
Transcript
CXorf64 has 1 exon which ultimately forms 1 transcript variant. This gene is expressed most in muscle tissue. There are no known isoforms in which CXorf64 is expressed.[4]
Protein
General Properties
| Property | Cleaved Protein | Mature Protein |
|---|---|---|
| Amino Acid Length | 298 | 298 |
| Isoelectric Point | 7.34 | ~7.2-7.4 |
| Molecular Weight | 31.9 kDa | ~31.9-33 kDa |
Structure
Shown to the right is a predicted tertiary structure of the protein. It is marked by long alpha-helices localized to the end of the protein opposite the N- and C- terminal ends

Expression
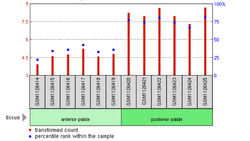
According to microarray-assessed tissue expression analysis by NCBI GEO, the gene CXorf64 has average expression levels in most tissues save for, prostate, and heart, fat, and endometrium which have relatively low levels of expression (~25th percentile of tissue gene expression) . Almost every microarray study determined very low levels of gene expression across all tissues assessed . This evidence prompted further analysis with other tools. While no tissue analysis was present on Allen Brain Atlas or genepaint, the Human Protein Atlas suggested that the gene was more highly expressed in heart, prostate, and a and fat tissues .
Sub-cellular Location
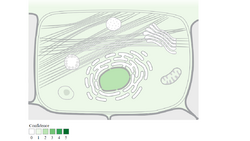
It synthesizes in the cell cytoplasm and is predicted to localize in the mitochondria. It is predicted to localize in the nucleus, plasma membrane, extracellular, mitochondrion, peroxisome, and cytosol.
Post-Translational Modification
N-linked glycosylation
It was found that there are several post-translational modifications that are performed on PRR32. These include several N-linked glycosylation sites that were predicted with high confidence. Glycosylation is known to play a part in cell-cell adhesion ( `a mechanism employed by cells of the immune system) via sugar-binding proteins. The graph illustrates predicted N-glyc sites across the protein chain (x-axis represents protein length from N- to C-terminal). A position with a potential (vertical lines) crossing the threshold (horizontal line at 0.5) is predicted glycosylated.[8]

Homology and Evolution
A list was generated of orthologs using the NCBI (protein) blast for PRR32, as well as UCSC’s BLAT (BLAST-Like Alignment Tool. These search and analyzing engines generated orthologs that are 80+long. This list was then narrowed down to a list of 30 different species. From those 30 species, they were divided into certain portions. The first to be selected were the primates since they are the closest related species when it comes to genetics. The similarity generated when compared to humans was approximately 95-99%. Then, using the edit and resubmit option on Blast (proteins), I excluded all primates in order to widen the species selection. The BLAST search again yielded many mammals including Rabbit, Dog, Ferret, Rhino, and many more. These species’ similarity percentage was anywhere between 69%-89%. Next, to further diversify my species selection, again using the same method, I excluded all mammals from the BLAST sequencing. This time, there were absolutely no matches in the inquiry. This action was repeated a few more times and it seems that the PRR32 gene is only relevant in mammals. The google spreadsheet created below lists the selection of species and all important information such as, order, year of divergence, etc. One thing to be noted is that the protein sequences for PRR32 were highly conserved amongst closely related species of Homo sapiens such as chimpanzees, gorillas and orangutans.
Clinical Significance
An experiment analyzed gene expression pattern in muscles from patients with amyotrophic lateral sclerosis (ALS) and multifocal motor neuropathy (MMN) compared to controls. Biopsied skeletal muscles from three ALS, three MMN and three control subjects had total RNA extracted and subjected to genome-wide gene expression analysis using Affymetrix GeneChip Exon 1.0 ST array. The most significant expression pattern differences were confirmed with RT-PCR in four additional ALS patients. Results showed that over 3000 genes were identified across the groups using q < 10%. Among 50 genes that were overexpressed only in the ALS group were: leucine-rich repeat kinase-2, follistatin, collagen type XIX alpha-1, ceramide kinase-like, sestrin-3 and CXorf64. No genes were significantly overexpressed in MMN alone. Underexpressed genes only in ALS included actinin αα3, fructose-1,6-bisphosphatase-2 and homeobox C10; whereas only in MMN: hemoglobin A1 and CXorf64. Ankyrin repeat domain-1 was overexpressed in both groups. Underexpressed genes in both groups included myosin light chain kinase-2, enolase-3 and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase-1. Validation analysis using RT-PCR confirmed the data for leucine-rich repeat kinase-2, follistatin, collagen type XIX alpha-1, ceramide kinase-like, sestrin-3 and CXorf64. In conclusion, there is differential tissue-specific gene expression in patients with ALS relative to MMN and controls. Further studies are necessary to evaluate the identified genes in larger patient groups and different tissues.
References
- ↑ "Entrez Gene: PRR32 proline rich 32". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=100130613.
- ↑ "Differential gene expression in patients with amyotrophic lateral sclerosis". Amyotrophic Lateral Sclerosis 12 (4): 250–6. July 2011. doi:10.3109/17482968.2011.560946. PMID 21375368.
- ↑ "PRR32 Gene - GeneCards | PRR32 Protein | PRR32 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=PRR32.
- ↑ "PRR32 proline rich 32 [Homo sapiens (human)] - Gene - NCBI". https://www.ncbi.nlm.nih.gov/gene/100130613.
- ↑ "PRR32 proline rich 32 [Homo sapiens (human)] - Gene - NCBI". https://www.ncbi.nlm.nih.gov/gene/100130613#.
- ↑ "Home - GEO - NCBI". https://www.ncbi.nlm.nih.gov/geo/.
- ↑ "Compartments - Prr32". http://compartments.jensenlab.org/Entity?figures=subcell_cell_%%&knowledge=10&textmining=10&predictions=10&type1=9606&type2=-22&id1=ENSP00000360166.
- ↑ "ExPASy: SIB Bioinformatics Resource Portal - Home". http://www.expasy.org/.
- ↑ "ExPASy: SIB Bioinformatics Resource Portal - Home". http://www.expasy.org/.
 |
