Biology:Reporter virus particles
Reporter virus particles (RVPs) are replication-incompetent virus particles engineered to express one or more reporter genes upon infecting susceptible cells.[1][2][3] Since the RVP genome lacks genes essential for viral replication, RVPs are capable of only a single round of infection.[4][5][6][7] Thus they are safe to work with under BSL-2 conditions, enabling the study of highly pathogenic viruses using standard laboratory facilities.[4][5][7][8][9][10] Expression of a reporter such as luciferase can provide a quantitative readout of infection.[4][5][10] With proper design and quality control, RVPs remain stable under common assay conditions and yield reproducible results that correlate with those obtained from live virus.[4][5] These qualities make RVPs a safer and faster alternative to plaque assays, and especially well-suited for high-throughput applications.[4][5][9][11][12] RVPs offer flexibility for different uses, as they are antigenically identical to wild-type virus, and can be engineered with various proteins or express mutant envelopes to study infectivity or antigenicity.[12][13]
Applications
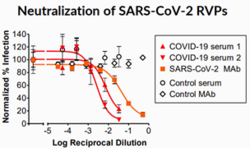
RVPs are most commonly used in neutralization assays, which measure the ability of serum or antibodies to prevent virus infectivity in vitro, with applications in vaccine development, antibody discovery, and serological testing.[14] A related assay tests for antibody-dependent enhancement (ADE), a phenomenon where non-neutralizing antibodies against viruses can increase infectivity through their binding to the cellular Fc receptor, aiding entry of the virus into host cells.[15]
Structure
Depending on the virus of interest and the desired application, RVPs can be pseudotypes, containing a heterologous self-assembling core (typically of lentiviral origin), as well as native envelope proteins corresponding to the studied virus.[2][3][16] This type of RVP facilitates exceptional reliability and reproducibility of neutralization assay results, while maintaining antigenicity and safety.[16] Alternatively, for structurally complex viruses such dengue and Zika viruses, RVPs are engineered to be antigenically identical to wild-type virus, using all of the structural proteins of the native virus.[1][4][5]
Limitations
RVP production requires optimization of several elements, such as expression constructs, cell lines, and processing steps, to reach a yield sufficient for downstream applications and reproducibility across production lots.[2] Although ideal for studying the effects of the immune response on virus entry, RVPs are replication-incompetent, and therefore typically do not allow study of the later stages of the viral life cycle. RVPs formed by pseudotyping contain the native form of the viral Envelope (or Spike) protein, but may not contain other structural elements from the original virus. Results obtained with RVPs are often compared to those obtained with live virus.[17]
References
- ↑ 1.0 1.1 Pierson, TC; Sánchez, MD; Puffer, BA; Ahmed, AA; Geiss, BJ; Valentine, LE; Altamura, LA; Diamond, MS; and Doms, RW (2006). "A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection". Virology. 346(1): 53-65.
- ↑ 2.0 2.1 2.2 King, B; Temperton, NJ; Grehan, K; Scott, SD; Wright, E; Tarr, AW; and Daly, JM (2016). "Technical considerations for the generation of novel pseudotyped viruses". Future Virology. 11(1): 47-59.
- ↑ 3.0 3.1 Li, Q; Liu, Q; Huang, W; Li, X; and Wang, Y (2018). "Current status on the development of pseudoviruses for enveloped viruses". Reviews in Medical Virology. 28(1): e1963.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Whitbeck, JC; Thomas, A; Kadash-Edmondson, K; Grinyo-Escuer, A; Stafford, LJ; Cheng, C; Liao, GC; Holtsberg, FW; Aman, MJ; Simmons, G; Davidson, E; and Doranz, BJ (2020). "Antigenicity, stability, and reproducibility of Zika reporter virus particles for long-term applications". PLoS Neglected Tropical Diseases. 14(11): e0008730.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Mattia, K; Puffer, BA; Williams, KL; Gonzalez, R; Murray, M; Sluzas, E; Pagano, D; Ajith, S; Bower, M; Berdougo, E; Harris, E; and Doranz, BJ (2011). "Dengue Reporter Virus Particles for measuring neutralizing antibodies against each of the four dengue serotypes". PLoS ONE. 6(11): e27252.
- ↑ Schmidt, F; Weisblum, Y; Muecksch, F; Hoffmann, H-H; Michailidis, E; Lorenzi, JCC; Mendoza, P; Rutkowska, M; Bednarski, E; Gaebler, C; Agudelo, M; Cho, A; Wang, Z; Gazumyan, A; Cipolla, M; Caskey, M; Robbiani, DF; Nussenzweig, MC; Rice, CM; Hatziioannou, T; and Bieniasz, PD (2020). "Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses". Journal of Experimental Medicine. 217(11).
- ↑ 7.0 7.1 Crawford, KHD; Eguia, R; Dingens, AS; Loes, AN; Malone, KD; Wolf, CR; Chu, HY; Tortorici, MA; Veesler, D; Murphy, M; Pettie, D; King, NP; Balazs, AB; and Bloom, JD (2020). "Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 Spike protein for neutralization assays". Viruses. 12(5): 513.
- ↑ Konduru, K; Shurtleff, AC; Bavari, S; and Kaplan, G (2018). "High degree of correlation between Ebola virus BSL-4 neutralization assays and pseudotyped VSV BSL-2 fluorescence reduction neutralization test". Journal of Virological Methods. 254: 1-7.
- ↑ 9.0 9.1 Peskova, M; Heger, Z; Janda, P; Adam, V; and Pekarik, V (2017). "An enzymatic assay based on luciferase Ebola virus-like particles for evaluation of virolytic activity of antimicrobial peptides". Peptides. 88: 87-96.
- ↑ 10.0 10.1 Tian, Y; Zhao, H; Liu, Q; Zhang, C; Nie, J; Huang, W; Li, C; Li, X; and Wang, Y (2018). "Development of in vitro and in vivo neutralization assays based on the pseudotyped H7N9 virus". Scientific Reports. 8(1): 8484.
- ↑ Xiao, JH; Rijal, P; Schimanski, L; Tharkeshwar, AK; Wright, E; Annaert, W; and Townsend, A (2018). "Characterization of Influenza Virus Pseudotyped with Ebolavirus Glycoprotein". Journal of Virology. 92(4): e00941-17.
- ↑ 12.0 12.1 Li, Q; Wu, J; Nie, J; Zhang, L; Hao, H; Liu, S; Zhao, C; Zhang, Q; Liu, H; Nie, L; Qin, H; Wang, M; Lu, Q; Li, X; Sun, Q; Liu, J; Zhang, L; Li, X; Huang, W; and Wang, Y (2020). "The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity". Cell. 182(5): 1284-1294.e9.
- ↑ Christian, EA; Kahle, KM; Mattia, K; Puffer, BA; Pfaff, JM; Miller, A; Paes, C; Davidson, E; and Doranz, BJ (2013). "Atomic-level functional model of dengue virus Envelope protein infectivity". Proceedings of the National Academy of Sciences. 110(46): 18662-18667.
- ↑ Ferrara, F and Temperton, N (2018). "Pseudotype neutralization assays: From laboratory bench to data analysis". Methods and Protocols. 1(1): 8.
- ↑ Valiant, WG and Mattapallil, JJ (2018). "A simple flow cytometry based assay to determine in vitro antibody dependent enhancement of dengue virus using Zika virus convalescent serum". Journal of Visualized Experiments(134).
- ↑ 16.0 16.1 Integral Molecular: SARS-CoV-2 Reporter Virus Particles. Available from: https://www.integralmolecular.com/wp-content/uploads/2020/07/INTG_SARS-CoV-2-RVPs_7_20.pdf.
- ↑ Xiang, Y; Nambulli, S; Xiao, Z; Liu, H; Sang, Z; Duprex, WP; Schneidman-Duhovny, D; Zhang, C; and Shi, Y (2020). "Versatile, multivalent nanobody cocktails efficiently neutralize SARS-CoV-2". bioRxiv: 2020.08.24.264333.
 |