Biology:ssNA-helicase RNA motif
| ssNA-helicase | |
|---|---|
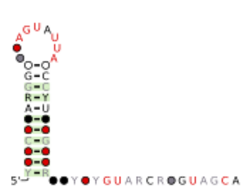 Consensus secondary structure and sequence conservation of ssNA-helicase RNA | |
| Identifiers | |
| Symbol | ssNA-helicase |
| Rfam | RF03070 |
| Other data | |
| RNA type | Gene; sRNA |
| SO | 0001263 |
| PDB structures | PDBe |
The ssNA-helicase RNA motif is a conserved RNA structure that was discovered by bioinformatics.[1] Although the ssNA-helicase motif was published as an RNA candidate, there is some reason to suspect that it might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available.
ssNA-helicase motif RNAs are found in Bacillota and Actinomycetota. The ssNA-helicase motif occurs upstream of genes that likely function as helicases, possible on an RNA substrate. Some evidence suggests that these genes are part of single-stranded RNA viruses (or presumably phages, as the RNAs are located in bacterial sequences). However, there is also a suggestion of an association with singled-stranded DNA viruses that infect plants. This latter potential association could mean that the ssNA-helicase motif actually functions as singled-stranded DNA. If the RNA is indeed part of a phage, then its location upstream of a protein-coding gene might just reflect the typical gene organization of phages, and not a direct functional or regulatory association between the RNA and the downstream gene. Therefore, it is ambiguous whether ssNA-helicase motif examples function as cis-regulatory elements or whether they operate in trans, and also whether they function as RNA or DNA.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |

