Biology:Tetraloop
From HandWiki
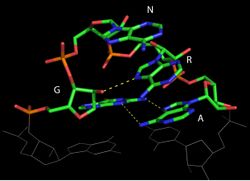
Tetraloops are a type of four-base hairpin loop motifs in RNA secondary structure that cap many double helices.[2] There are many variants of the tetraloop. The published ones include ANYA,[3][4] CUYG,[5] GNRA,[6] UNAC[7] and UNCG.[8]
Three types of tetraloops are common in ribosomal RNA: GNRA, UNCG and CUUG, in which the N could be either uracil, adenine, cytosine, or guanine, and the R is either guanine or adenine. These three sequences form stable and conserved tetraloops that play an important role in structural stability and biological function of 16S rRNA.[9]
- GNRA
- The GNRA tetraloop has a guanine-adenine base-pair where the guanine is 5' to the helix and the adenine is 3' to the helix. Tetraloops with the sequence UMAC have essentially the same backbone fold as the GNRA tetraloop,[7] but may be less likely to form tetraloop-receptor interactions. They may therefore be a better choice for closing stems when designing artificial RNAs.
- The presence of the GNRA tetraloop provides an exceptional stability to RNA structure. GNRA occurs 50% more than other tetranucleotides due to their ability to withstand temperatures 4 °C higher than other RNA hairpins. This allows them to act as nucleation sites for proper folding of RNA. The rare hydrogen bonds between the first guanine and fourth adenine nucleotide, extensive stacking of nucleotide bases and hydrogen bonds between 2' OH of a ribose sugar and nitrogenous bases makes the tetraloop thermodynamically stable.[10]
- UNCG
- In the UNCG is favorable thermodynamically and structurally due to hydrogen bonding, van der Waals interactions, coulombic interactions and the interactions between the RNA and the solvent. The UNCG tetraloops are more stable than DNA loops with the same sequence. The UUCG tetraloop is the most stable tetraloop.[11] UUCG and GNRA tetraloops make up 70% of all tetraloops in 16S-rRNA .[2]
- CUUG
See also
- RNA tertiary structure (section Tetraloop-receptor interactions)
References
- ↑ Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., Doudna, J.A. (1996). "Crystal structure of a group I ribozyme domain: principles of RNA packing.". Science 273 (5282): 1676–1685. doi:10.1126/science.273.5282.1678. PMID 8781224. Bibcode: 1996Sci...273.1678C.
- ↑ 2.0 2.1 Woese, C.R., Winkers, S., Gutell, R.R. (1990). "Architecture of ribosomal RNA: Constraints on the sequence of "tetra-loops"". Proc. Natl. Acad. Sci. USA 87 (21): 8467–71. doi:10.1073/pnas.87.21.8467. PMID 2236056. Bibcode: 1990PNAS...87.8467W.
- ↑ Zirbel, CL; Sponer, JE; Sponer, J; Stombaugh, J; Leontis, NB (Aug 2009). "Classification and energetics of the base-phosphate interactions in RNA.". Nucleic Acids Research 37 (15): 4898–918. doi:10.1093/nar/gkp468. PMID 19528080.
- ↑ Klosterman, PS; Hendrix, DK; Tamura, M; Holbrook, SR; Brenner, SE (2004). "Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns.". Nucleic Acids Research 32 (8): 2342–52. doi:10.1093/nar/gkh537. PMID 15121895.
- ↑ Jucker, FM; Pardi, A (Nov 7, 1995). "Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif.". Biochemistry 34 (44): 14416–27. doi:10.1021/bi00044a019. PMID 7578046.
- ↑ Jaeger, L; Michel, F; Westhof, E (Mar 11, 1994). "Involvement of a GNRA tetraloop in long-range RNA tertiary interactions.". Journal of Molecular Biology 236 (5): 1271–6. doi:10.1016/0022-2836(94)90055-8. PMID 7510342.
- ↑ 7.0 7.1 Zhao, Q; Huang, HC; Nagaswamy, U; Xia, Y; Gao, X; Fox, GE (Aug 2012). "UNAC tetraloops: to what extent do they mimic GNRA tetraloops?". Biopolymers 97 (8): 617–28. doi:10.1002/bip.22049. PMID 22605553.
- ↑ Molinaro, M; Tinoco I, Jr (Aug 11, 1995). "Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications.". Nucleic Acids Research 23 (15): 3056–63. doi:10.1093/nar/23.15.3056. PMID 7544890.
- ↑ 9.0 9.1 Baumruk, Vladimir; Gouyette, Catherine; Huynh-Dinh, Tam; Sun, Jian-Sheng; Ghomi, Mahmoud (2001-10-01). "Comparison between CUUG and UUCG tetraloops: thermodynamic stability and structural features analyzed by UV absorption and vibrational spectroscopy". Nucleic Acids Research 29 (19): 4089–4096. doi:10.1093/nar/29.19.4089. ISSN 0305-1048. PMID 11574692.
- ↑ Heus, Hans A.; Pardi, Arthur (1991-01-01). "Structural Features that Give Rise to the Unusual Stability of RNA Hairpins Containing GNRA Loops". Science 253 (5016): 191–194. doi:10.1126/science.1712983. PMID 1712983. Bibcode: 1991Sci...253..191H.
- ↑ Antao, V. P.; Lai, S. Y.; Tinoco, I. (1991-11-11). "A thermodynamic study of unusually stable RNA and DNA hairpins". Nucleic Acids Research 19 (21): 5901–5905. doi:10.1093/nar/19.21.5901. ISSN 0305-1048. PMID 1719483.
- ↑ Hall, Kathleen B. (October 15, 2013). "RNA does the folding dance of twist, turn, stack". Proceedings of the National Academy of Sciences of the United States of America 110 (42): 16706–7. doi:10.1073/pnas.1316029110. PMID 24072647. Bibcode: 2013PNAS..11016706H.
 |
