Biology:Yeast U1 spliceosomal RNA
From HandWiki
| Yeast U1 spliceosomal RNA | |
|---|---|
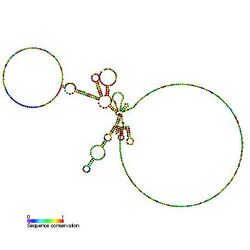 Predicted secondary structure and sequence conservation of U1_yeast | |
| Identifiers | |
| Symbol | U1_yeast |
| Rfam | RF00488 |
| Other data | |
| RNA type | Gene; snRNA; splicing |
| Domain(s) | Eukaryota |
| GO | 0000368 0030627 0005685 |
| SO | 0000391 |
| PDB structures | PDBe |
U1 is a small nuclear RNA (snRNA) component of the spliceosome and is involved in pre-mRNA splicing.
In the splicing process the 5' end of the U1 snRNA forms complementary base pairing with the 5' splice junction of the intron to be excised, thus defining the 5' donor site of an intron.
There are significant differences in sequence and secondary structure between metazoan and yeast U1 snRNAs, the latter being much longer (568 nucleotides as compared to 164 nucleotides in human). Nevertheless, secondary structure predictions suggest that all U1 snRNAs share a 'common core' consisting of helices I, II, the proximal region of III, and IV.[1][2] The secondary structure model shows the structure prediction for the larger yeast sequences.
References
- ↑ Zwieb C (1997). "The uRNA database". Nucleic Acids Res. 25 (1): 102–3. doi:10.1093/nar/25.1.102. PMID 9016512.
- ↑ "Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains". Proc. Natl. Acad. Sci. U.S.A. 87 (2): 851–5. 1990. doi:10.1073/pnas.87.2.851. PMID 2405391. Bibcode: 1990PNAS...87..851K.
External links
 |
