Chemistry:Abductin
Abductin is a naturally occurring elastomeric protein found in the hinge ligament of bivalve mollusks. It is unique as it is the only natural elastomer with compressible elasticity, as compared to resilin, spider silk, and elastin.[1] Its name was proposed from the fact that it functions as the abductor of the valves of bivalve mollusks.
The properties of abductin vary across species of bivalves due to the specific use case of the species or the environment the species is found in. In spite of these differences, the same general function of acting opposite of the abductor muscles, where the resilin forces the shells into an open configuration.
Though patents for a specific protein sequences of abductin were approved by the United States Patent and Trademark Offices, there are no large scale commercial uses for abductin as of April 2022.
Structure
Amino acid composition
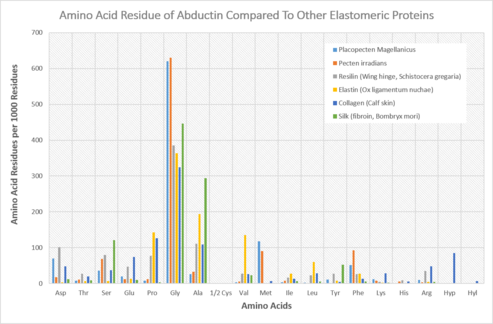
The amino acid composition of protein within the inner hinge ligament of bivalve mollusks was first discovered by Robert E. Kelly and Robert V. Rice in 1967, who subsequently proposed the protein’s name as abductin.[3] This was derived from its function as the abductor of the shells of bivalve mollusks. Kelly and Rice discovered that the protein lacked the presence of hydroxyproline and hydroxylysine, which are amino acids indicative of the common protein, collagen. Further analysis showed that abductin is made of three prominent amino acids: glycine, methionine, and phenylalanine, which are arranged in multiple repeating sequences throughout the molecule.[4] This was found in Placopecten magellanicus. Abductin is similar to elastin and resilin, but has a main difference having high concentrations of glycine and methionine.[3] The glycine and methionine, and other amino acid residues, vary in concentration with different species. In Argopecten irradians, for example, glycine and methionine make up 57.3% and 14.3% of the protein, respectively.[4] The high concentration of methionine found in abductin makes it unique because it is not a common occurrence in natural elastomeric proteins.
Protein structure

Peptide sequences such as MGGG, FGGMG, FGGMGGG, GGFGGMGGG, and FGGMGGGNAG are repeated throughout the peptide chain.[3] It is to note that these peptide sequences all contain glycine. Additionally, in Argopecten irradians, the pentapeptide FGGMG is repeated throughout the molecule.[5] The main peptide sequence feature of abductin is the presence of many repeating sequences, all of which contain glycine residues. This is similar to that of the structure of elastin.
Abductin is lightly cross-linked, which gives it its high elasticity. The source of cross-linking has been researched, but no concrete explanation has been devised. The lack of tyrosine in the peptide chain suggests that cross-links are not formed through dityrosine links, like it is in resilin.[5] Hypotheses of the mechanism of cross-linking have been proposed by various researchers. One potential source of cross-linking is due to the presence of a methionine dimer, ½ cystine in some species, or other similar amino acids that contains a disulfide bridge, which creates the cross-link between peptide chains.[2][3] Another study discovered that 3,3'-methylene-bistyrosine could be responsible for the cross-linkage in abductin, similar to how tyrosine and lysine residues are responsible for the cross-linking in resilin and elastin.[6]
Abductin is acellular and amorphous in structure, as discovered through microscopy and x-ray diffraction, respectively.[2] Since abductin is insoluble and its isolation from the hinge ligament is difficult, there is a lack of research concerning its structure at the protein level, such as secondary and hierarchical structures.[2] More recent research on synthetic peptides derived from abductin were found to have polyproline II helix structure in aqueous solutions and type II β-turn structure in hydrophobic solvents.[1] Combinations of both structures can also be observed for longer abductin-like peptide chains.[1]
Biological function
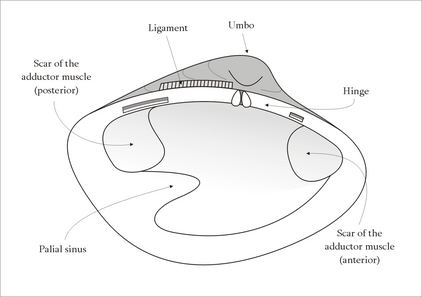
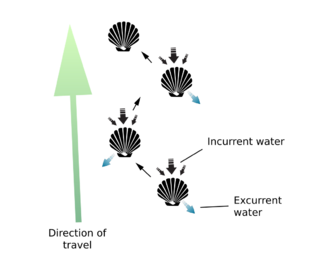
The use of abductin varies among the different species of mollusks in the world. Some, like scallops and file shells, are able to swim using a repetitive motion of opening and closing its shell, the motion of which rapidly intakes and expels water.[7] In other species of mollusks, the presence of abductin is usually located where the two shells come together to form a hinge.[8] Unlike the needs of scallops for efficient energy return for the purpose of movement, species like the Apylsia find it necessary to reduce energy return in favor of stability in opening and closing of the shells.[8] Abductin can be found within the resilium structure, which is used to store mechanical energy for this purpose.[7] The effectiveness of abductin is highly influenced by the morphological aspects of the mollusk's shell, such as its size and shape.[7] Other influences on the performance of abductin in mollusks is temperature, where there is a decrease in performance as the temperature of the surrounding environment decreases, and the presence of octopine - which acts as an analogous to lactic acid in mammals.[7] The implementation of the resilium structure of the clam can be modeled as an oscillatory system, where it works against the abductor muscle to open the shell of the organism; the resilium forces the shell open while the abductor muscle control the shell’s closure.[7]
Material properties
Little data exist on the structure and function of compressible elastomeric proteins such as abductin. An understanding of the underlying structural features of these proteins may lead to the development of a new class of highly tailored ‘‘compressible’’ hydrogels. Gaining knowledge of the underlying structural and functional features of compressible natural elastomers, such as abductin, can lead to novel compressible bioelastomers with tailored material properties.[9]
Solubility
By interpreting Hurst exponents as Flory, water results to be a poor solvent for the abductin peptides.[10] Predicting the functional solvent environment for insoluble proteins like abductin is particularly difficult because the protein’s hydrophobicity and the probable cross-linked nature suggest a less polar internal environment than the surrounding solvent.[9]
Conformation
The presence of both extended conformations (PPII) and folded conformations (β-turns) in equilibrium to describe abductin has been previously suggested.[9] Circular Dichroism (CD) spectra revealed that AMP1 (a 25 amino acid abductin sequence) adopts a dominant unordered conformation at 258 °C and a polyproline II (PPII) conformation at 0 °C and 458 °C with a possible minor amount of type II β-turn conformers.[9] This observation indicates that AMP1 undergoes an inverse temperature transition in that it goes from a dominant unordered conformation to a periodic, extended PPII conformation with increasing temperature.[9] The secondary structure of abductin was also investigated by Nuclear Magnetic Resonance (NMR) and CD studies of several synthetic peptides. Most synthetic abductin-based peptides adopted polyproline II (PPII) structures, which are left-handed helices, in aqueous solution, whereas they had type II β-turns in trifluoroethanol (TFE), which is a more hydrophobic (less polar) solvent. The coexistence of PPII and type II β-turns and temperature-induced multiconformational transitions were observed with longer synthetic abductin-like peptides such as (FGGMGGGNAG)4 in hexafluoroisopropanol (HFIP).[11] The secondary structure of AB12 was qualitatively analyzed by comparing the CD spectra to other peptides with known secondary structures. The CD spectra of aqueous solutions of AB12 shows a strong negative peak at 200 nm and a tendency toward positive values at ~218 nm, which are characteristics of PPII helices. An isodichroic point at ~208 nm suggests an equilibrium exists between the PPII structure and other conformations. In addition, because the peak at 218 nm never exceeds zero, the spectra suggest the coexistence of unordered structures and PPII helices. A small negative band can be observed at ~225 nm, which likely results from the aromatic residue, phenylalanine, in the sequence.[11]
Temperature
The effect of temperature on the secondary structure was studied. With increasing temperature, the magnitude of both peaks in the CD spectra at 200 and 218 nm decreased, which is typical for PPII helix conformations. In addition, the change in structure because of temperature was fully reversible and did not display any hysteresis. The PPII conformation, which is widely present in elastomeric proteins such as elastin and titin, is believed to play an important role in determining the elasticity of these proteins.[11] The abductin-based protein possessed reversible Upper Critical Solution Temperature (UCST) behavior and formed a gel-like structure. At high temperatures, it displayed irreversible aggregation behavior. Thermal responsiveness is a useful property for engineering drug delivery systems because the encapsulation and release of drugs can easily be controlled via temperature change.[11]
Cytocompatibility
The abductin-based protein was cytocompatible, and cells spread slowly when first seeded on the abductin-based protein.[11] A LIVE/DEAD assay revealed that human umbilical vein endothelial cells had a viability of 98 ± 4% after being cultured for two days on the abductin-based protein. Initial cell spreading on the abductin-based protein was similar to that on bovine serum albumin. These studies thus demonstrate the potential of abductin-based proteins in tissue engineering and drug delivery applications due to the cytocompatibility and its response to temperature.[11]
Tensile and compressive moduli
Natural abductin has a tensile modulus of 1.25 MPa, which is higher than elastin (0.3−0.6 MPa) but on the same order of magnitude as resilin (0.6−2 MPa).[11] It has a compressive modulus of 4 MPa, which is higher than resilin (0.6−0.7 MPa). The superior mechanical properties of natural abductin offer the potential for designing protein-based biomaterials that can be utilized in a broader number of applications.[11]
Hydrodynamic volume and temperature relationship
A solution of AB12 (10 mg/mL in Milli-Q water) was visually observed to turn from transparent to opaque when cooled from room temperature to lower temperatures (incubated on ice). Dynamic Light Scattering (DLS) was used to further investigate the temperature responsiveness of AB12. An abrupt decrease in the hydrodynamic diameter (DH) of AB12 was observed when the protein solution was heated from 2 to 5 °C. This phenomenon is indicative of Upper Critical Solution Temperature (UCST) behavior. The change in DH at low temperatures was reversible and displayed some hysteresis. A moderate increase in DH was observed from 35 °C, and a sharper increase in DH occurred starting at 57 °C (aggregation temperature). Compared to the reversible UCST behavior, the transition that occurred at the aggregation temperature was irreversible.[11]
Extended-folded behavior
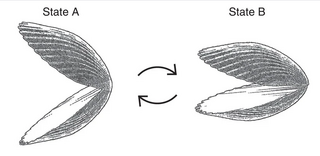
In the case of abductin, on compression, the equilibrium extended ⇄ folded should be shifted to the folded structures, decreasing the entropy. The uncompressed, multi-conformational state is recovered by a simple increase in entropy after the removal of the compression force. This is opposite to elastin’s behavior.[12]
Engineering applications
The first patent that is dedicated to the usage and implementation of abductin was accepted by the United States Patent and Trademark Office on October 3, 2000 (Patent No. 6,127,166).[13] The patent in question details the specific protein sequence of abductin to be manufactured through biological means and the possible applications of the polymer, suggesting possible uses as a copolymer for other naturally occurring polymers, a fabric material, or a material that binds with antibodies.[13] As of April 2022, there hasn’t been large-scale production, nor application, of polymers derived from the abductin or related polymeric sequences.
References
- ↑ 1.0 1.1 1.2 R. S.-C. Su, J. N. Renner, and J. C. Liu, "Synthesis and characterization of recombinant abductin-based proteins", Biomacromolecules, vol. 14, no. 12, pp. 4301–4308, 2013.
- ↑ 2.0 2.1 2.2 2.3 George A. Kahler, Frank M. Fisher, and Ronald L. Sass, "The chemical composition and mechanical properties of the hinge ligament in bivalve molluscs", The Biological Bulletin, vol. 151, no. 1, pp. 161–181, 1976.
- ↑ 3.0 3.1 3.2 3.3 R. E. Kelly and R. V. Rice, "Abductin: A rubber-like protein from the internal triangular hinge ligament of pecten", Science, vol. 155, no. 3759, pp. 208–210, 1967.
- ↑ 4.0 4.1 4.2 Q. Cao, Y. Wang, and H. Bayley, "Sequence of abductin, the molluscan 'rubber' protein", Current Biology, vol. 7, no. 11, 1997.
- ↑ 5.0 5.1 H. Ehrlich, "Chapter 19: Abductin", in Biological Materials of Marine Origin: Vertebrates, Springer.
- ↑ S. O. Andersen, "Isolation of a new type of cross link from the hinge ligament protein of molluscs", Nature, vol. 216, no. 5119, pp. 1029–1030, 1967
- ↑ 7.0 7.1 7.2 7.3 7.4 Mark Denny, Luke Miller; "Jet propulsion in the cold: mechanics of swimming in the Antarctic scallop Adamussium colbecki". Journal of Experimental Biology, 15 November 2006; 209 (22): 4503–4514.
- ↑ 8.0 8.1 Sutton, G.P., Macknin, J.B., Gartman, S.S. et al. "Passive hinge forces in the feeding apparatus of Aplysia aid retraction during biting but not during swallowing". Journal of Comparative Physiology A, 190, 501–514 (2004).
- ↑ 9.0 9.1 9.2 9.3 9.4 Bochicchio, B., Jimenez-Oronoz, F., Pepe, A., Blanco, M., Sandberg, L. and Tamburro, A., 2005. "Synthesis of and Structural Studies on Repeating Sequences of Abductin". Macromolecular Bioscience, [online] 5(6), pp.502-511
- ↑ Villani, V., 2003. "Complexity of polypeptide dynamics: chaos, Brownian motion and elasticity in aqueous solution". Journal of Molecular Structure: THEOCHEM, [online] 621(1-2), pp.127-139.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 11.8 Bochicchio, B., Pepe, A. and Tamburro, A., 2005. "Circular dichroism studies on repeating polypeptide sequences of abductin". Chirality, [online] 17(7), pp.364-372.
- ↑ Su, R., Renner, J. and Liu, J., 2013. "Synthesis and Characterization of Recombinant Abductin-Based Proteins". Biomacromolecules, [online] 14(12), pp.4301-4308.
- ↑ 13.0 13.1 U.S. Patent 6,127,166
 |