Chemistry:Fluorescent D-amino acids
Fluorescent D-amino acids (FDAAs) are D-amino acid derivatives whose side-chain terminal is covalently coupled with a fluorophore molecule.[1] FDAAs incorporate into the bacterial peptidoglycan (PG) in live bacteria, resulting in strong peripheral and septal PG labeling without affecting cell growth. They are featured with their in-situ incorporation mechanisms which enable time-course tracking of new PG formation.[2] To date, FDAAs have been employed for studying the cell wall synthesis in various bacterial species (both Gram-positives and Gram-negatives) through different techniques, such as microscopy, mass spectrometry, flow cytometry.
Structures and general properties
FDAA consists of a D-amino acid and a fluorophore (coupled through the amino acid side chain.) The D-amino acid backbone is required for its incorporation into the bacterial peptidoglycan through the activity of DD-transpeptidases.[3] Once being incorporated, one can use fluorescence-detection techniques to visualize the location of new PG formation as well as the growth rate.[4]
D-alanine is the most well-studied D-amino acids for FDAA development because it is a naturally existing residue in bacterial peptidoglycan structures. On the other hand, various fluorophores have been employed for FDAA applications and each has its features.[5] For example, coumarin-based FDAA (HADA) is small enough to penetrate the bacterial outer-membranes and thus is widely used for Gram-negative bacterial studies; while TAMRA-based FDAA (TADA) features its high brightness and photo/thermo-stability, which is suitable for super-resolution microscopy (strong excitation light is used).[5]
Proposed FDAA incorporation mechanisms
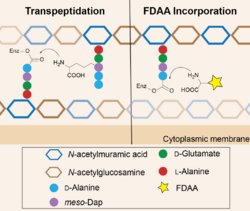
Peptidoglycan (PG) is a mesh-like structure containing polysaccharides cross-linked by peptide chains.[6] Penicillin-binding proteins (DD-transpeptidases), in short PBPs, recognize the PG peptides and catalyze the cross-linking reactions.[7] These enzymes are reported to have high specificity toward the chirality center of the amino acid backbone (the D-chiral center) but relatively low specificity toward the side-chain structure. Therefore, when FDAAs are present, they are taken by PBPs for the cross-linking reactions, resulting in their incorporation into the PG peptide chains. At proper concentration, e.g. 1-2 mM, FDAAs labeling does not affect PG synthesis and cell growth because only 1-2% of PG peptide chains are labeled with FDAA.[2] File:Sequential labeling of S venezuelae.tif
Applications
Published studies utilizing FDAAs as tools include:
- Visualizing bacterial cell wall structures.[2]
- Studying bacterial cell wall growth.[1][4]
- Monitoring bacterial cell wall turnover.[8][9]
- Quantifying bacterial cell wall growth activity.[10]
- Assaying the anti-cell wall ability of antibiotics.[1]
- Screening new anti-cell wall antibiotics.[11]
- Tracking transpeptidase activity in vitro.[12]
References
- ↑ Jump up to: 1.0 1.1 1.2 1.3 Hsu, Yen-Pang; Booher, Garrett; Egan, Alexander; Vollmer, Waldemar; VanNieuwenhze, Michael S. (2019-09-17). "d -Amino Acid Derivatives as in Situ Probes for Visualizing Bacterial Peptidoglycan Biosynthesis" (in en). Accounts of Chemical Research 52 (9): 2713–2722. doi:10.1021/acs.accounts.9b00311. ISSN 0001-4842. PMID 31419110. https://pubs.acs.org/doi/10.1021/acs.accounts.9b00311.
- ↑ Jump up to: 2.0 2.1 2.2 Kuru, Erkin; Hughes, H. Velocity; Brown, Pamela J.; Hall, Edward; Tekkam, Srinivas; Cava, Felipe; de Pedro, Miguel A.; Brun, Yves V. et al. (2012-12-07). "In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D -Amino Acids" (in en). Angewandte Chemie International Edition 51 (50): 12519–12523. doi:10.1002/anie.201206749. PMID 23055266.
- ↑ Kuru, Erkin; Radkov, Atanas; Meng, Xin; Egan, Alexander; Alvarez, Laura; Dowson, Amanda; Booher, Garrett; Breukink, Eefjan et al. (2019-12-20). "Mechanisms of Incorporation for D -Amino Acid Probes That Target Peptidoglycan Biosynthesis" (in en). ACS Chemical Biology 14 (12): 2745–2756. doi:10.1021/acschembio.9b00664. ISSN 1554-8929. PMID 31743648.
- ↑ Jump up to: 4.0 4.1 Radkov, Atanas D.; Hsu, Yen-Pang; Booher, Garrett; VanNieuwenhze, Michael S. (2018-06-20). "Imaging Bacterial Cell Wall Biosynthesis" (in en). Annual Review of Biochemistry 87 (1): 991–1014. doi:10.1146/annurev-biochem-062917-012921. ISSN 0066-4154. PMID 29596002.
- ↑ Jump up to: 5.0 5.1 Hsu, Yen-Pang; Rittichier, Jonathan; Kuru, Erkin; Yablonowski, Jacob; Pasciak, Erick; Tekkam, Srinivas; Hall, Edward; Murphy, Brennan et al. (2017). "Full color palette of fluorescent d -amino acids for in situ labeling of bacterial cell walls" (in en). Chemical Science 8 (9): 6313–6321. doi:10.1039/C7SC01800B. ISSN 2041-6520. PMID 28989665. PMC 5628581. http://xlink.rsc.org/?DOI=C7SC01800B.
- ↑ Vollmer, Waldemar; Blanot, Didier; De Pedro, Miguel A. (March 2008). "Peptidoglycan structure and architecture" (in en). FEMS Microbiology Reviews 32 (2): 149–167. doi:10.1111/j.1574-6976.2007.00094.x. ISSN 1574-6976. PMID 18194336.
- ↑ Typas, Athanasios; Banzhaf, Manuel; Gross, Carol A.; Vollmer, Waldemar (February 2012). "From the regulation of peptidoglycan synthesis to bacterial growth and morphology" (in en). Nature Reviews Microbiology 10 (2): 123–136. doi:10.1038/nrmicro2677. ISSN 1740-1526. PMID 22203377.
- ↑ Boersma, Michael J.; Kuru, Erkin; Rittichier, Jonathan T.; VanNieuwenhze, Michael S.; Brun, Yves V.; Winkler, Malcolm E. (2015-11-01). de Boer, P.. ed. "Minimal Peptidoglycan (PG) Turnover in Wild-Type and PG Hydrolase and Cell Division Mutants of Streptococcus pneumoniae D39 Growing Planktonically and in Host-Relevant Biofilms" (in en). Journal of Bacteriology 197 (21): 3472–3485. doi:10.1128/JB.00541-15. ISSN 0021-9193. PMID 26303829.
- ↑ Kuru, Erkin; Lambert, Carey; Rittichier, Jonathan; Till, Rob; Ducret, Adrien; Derouaux, Adeline; Gray, Joe; Biboy, Jacob et al. (December 2017). "Fluorescent D-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation" (in en). Nature Microbiology 2 (12): 1648–1657. doi:10.1038/s41564-017-0029-y. ISSN 2058-5276. PMID 28974693.
- ↑ Bisson-Filho, Alexandre W.; Hsu, Yen-Pang; Squyres, Georgia R.; Kuru, Erkin; Wu, Fabai; Jukes, Calum; Sun, Yingjie; Dekker, Cees et al. (2017-02-17). "Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division" (in en). Science 355 (6326): 739–743. doi:10.1126/science.aak9973. ISSN 0036-8075. PMID 28209898. Bibcode: 2017Sci...355..739B.
- ↑ Culp, Elizabeth J.; Waglechner, Nicholas; Wang, Wenliang; Fiebig-Comyn, Aline A.; Hsu, Yen-Pang; Koteva, Kalinka; Sychantha, David; Coombes, Brian K. et al. (2020-02-27). "Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling" (in en). Nature 578 (7796): 582–587. doi:10.1038/s41586-020-1990-9. ISSN 0028-0836. PMID 32051588. Bibcode: 2020Natur.578..582C. http://www.nature.com/articles/s41586-020-1990-9.
- ↑ Hsu, Yen-Pang; Hall, Edward; Booher, Garrett; Murphy, Brennan; Radkov, Atanas D.; Yablonowski, Jacob; Mulcahey, Caitlyn; Alvarez, Laura et al. (April 2019). "Fluorogenic d-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays" (in en). Nature Chemistry 11 (4): 335–341. doi:10.1038/s41557-019-0217-x. ISSN 1755-4330. PMID 30804500. Bibcode: 2019NatCh..11..335H.
External links
 |


