Physics:Random coil index
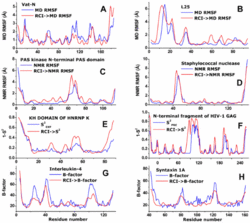
Random coil index (RCI) predicts protein flexibility by calculating an inverse weighted average of backbone secondary chemical shifts and predicting values of model-free order parameters as well as per-residue RMSD of NMR and molecular dynamics ensembles from this parameter.[1]
The key advantages of this protocol over existing methods of studying protein flexibility are
- it does not require prior knowledge of a protein's tertiary structure,
- it is not sensitive to the protein's overall tumbling and
- it does not require additional NMR measurements beyond the standard experiments for backbone assignments.[2]
The application of secondary chemical shifts to characterize protein flexibility is based on an assumption that the proximity of chemical shifts to random coil values is a manifestation of increased protein mobility, while significant differences from random coil values are an indication of a relatively rigid structure.[1]
Even though chemical shifts of rigid residues may adopt random coil values as a result of comparable contributions of shielding and deshielding effects (e.g. from torsion angles, hydrogen bonds, ring currents, etc.), combining the chemical shifts from multiple nuclei into a single parameter allows one to decrease the influence of these flexibility false positives. The improved performance originates from the different probabilities of random coil chemical shifts from different nuclei being found among amino acid residues in flexible regions versus rigid regions. Typically, residues in rigid helices or rigid beta-strands are less likely to have more than one random coil chemical shift among their backbone shifts than residues in mobile regions.[2]
The actual calculation of the RCI involves several additional steps including the smoothing of secondary shifts over several adjacent residues, the use of neighboring residue corrections, chemical shift re-referencing, gap filling, chemical shift scaling and numeric adjustments to prevent divide-by-zero problems. 13C, 15 N and 1H secondary chemical shifts are then scaled to account for the characteristic resonance frequencies of these nuclei and to provide numeric consistency among different parts of the protocol. Once these scaling corrections have been done, the RCI is calculated. The ‘‘end-effect correction’’ can also be applied at this point. The last step of the protocol involves smoothing the initial set of RCI values by three-point averaging.[3] [4]
See also
- Chemical Shift
- Chemical shift index
- Protein dynamics
- Protein NMR
- NMR
- Nuclear magnetic resonance spectroscopy
- Protein nuclear magnetic resonance spectroscopy
- Protein dynamics
- Protein
References
- ↑ 1.0 1.1 Mark, Berjanskii; David Wishart (2005). "A simple method to predict protein flexibility using secondary chemical shifts". Journal of the American Chemical Society 127 (43): 14970–14971. doi:10.1021/ja054842f. PMID 16248604.
- ↑ 2.0 2.1 Mark, Berjanskii; David Wishart (2008). "Application of the random coil index to studying protein flexibility". Journal of Biomolecular NMR 40 (1): 31–48. doi:10.1007/s10858-007-9208-0. PMID 17985196.
- ↑ Mark, Berjanskii; David Wishart (2006). "NMR: prediction of protein flexibility". Nature Protocols 1 (2): 683–688. doi:10.1038/nprot.2006.108. PMID 17406296.
- ↑ Mark, Berjanskii; David Wishart (2007). "The RCI server: rapid and accurate calculation of protein flexibility using chemical shifts". Nucleic Acids Research 35 (Web Server issue): W531–W537. doi:10.1093/nar/gkm328. PMID 17485469.
 |
