Biology:Streptavidin
| Avidin family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
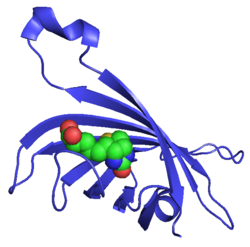 Monomeric streptavidin (ribbon diagram) with bound biotin (spheres) | |||||||||
| Identifiers | |||||||||
| Symbol | Avidin | ||||||||
| Pfam | PF01382 | ||||||||
| InterPro | IPR005468 | ||||||||
| PROSITE | PDOC00499 | ||||||||
| SCOP2 | 1slf / SCOPe / SUPFAM | ||||||||
| |||||||||
Streptavidin /ˌstrɛpˈtævɪdɪn/ is a 52.8 kDa protein purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin H). With a dissociation constant (Kd) on the order of ≈10−14 mol/L,[1] the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants (e.g. guanidinium chloride), detergents (e.g. SDS, Triton), proteolytic enzymes, and extremes of temperature and pH.
Structure
The crystal structure of streptavidin with biotin bound was reported by two groups in 1989. The structure was solved using multi wavelength anomalous diffraction by Hendrickson et al.[2] at Columbia University and using multiple isomorphous replacement by Weber et al.[3] at E. I. DuPont Central Research and Development Department. As of September 2017, there are 171 structures deposited in the Protein Data Bank. See this link for a complete list. The N and C termini of the 159 residue full-length protein are processed to give a shorter ‘core’ streptavidin, usually composed of residues 13 - 139; removal of the N and C termini is necessary for the high biotin-binding affinity. The secondary structure of a streptavidin monomer is composed of eight antiparallel β-strands, which fold to give an antiparallel beta barrel tertiary structure. A biotin binding-site is located at one end of each β-barrel. Four identical streptavidin monomers (i.e. four identical β-barrels) associate to give streptavidin’s tetrameric quaternary structure. The biotin binding-site in each barrel consists of residues from the interior of the barrel, together with a conserved Trp120 from neighboring subunit. In this way, each subunit contributes to the binding site on the neighboring subunit, and so the tetramer can also be considered a dimer of functional dimers.
Origins of the high affinity
The numerous crystal structures of the streptavidin-biotin complex have shed light on the origins of the remarkable affinity. Firstly, there is high shape complementarity between the binding pocket and biotin. Secondly, there is an extensive network of hydrogen bonds formed to biotin when in the binding site. There are eight hydrogen bonds directly made to residues in the binding site (the so-called 'first shell' of hydrogen bonding), involving residues Asn23, Tyr43, Ser27, Ser45, Asn49, Ser88, Thr90 and Asp128. There is also a 'second shell' of hydrogen bonding involving residues that interact with the first shell residues. However, the streptavidin-biotin affinity exceeds that which would be predicted from the hydrogen bonding interactions alone, suggesting another mechanism contributing to the high affinity.[4] The biotin-binding pocket is hydrophobic, and there are numerous van der Waals force-mediated contacts and hydrophobic interactions made to the biotin when in the pocket, which is also thought to account for the high affinity. In particular, the pocket is lined with conserved tryptophan residues. Lastly, biotin binding is accompanied by the stabilisation of a flexible loop connecting B strands 3 and 4 (L3/4), which closes over the bound biotin, acting like a 'lid' over the binding pocket and contributing to the extremely slow biotin dissociation rate.
Most attempts at mutating streptavidin result in a lowered biotin-binding affinity, which is to be expected in such a highly optimised system. However, a recently engineered mutant of streptavidin, named traptavidin, was found to have more than ten-fold slower biotin dissociation, in addition to higher thermal and mechanical stability,.[5] This decreased dissociation rate was accompanied by a two-fold decrease in the association rate.
Biotin-binding affinity can be impaired by chemical labeling of streptavidin, such as with amine-reactive fluorophores. Flavidin is a streptavidin mutant without lysine side-chains, which retains good biotin binding characteristics after such fluorescent dye labeling.[6]
Uses in Biotechnology
Among the most common uses are the purification or detection of various biomolecules. The strong streptavidin-biotin bond can be used to attach various biomolecules to one another or onto a solid support. Harsh conditions are needed to break the streptavidin-biotin interaction, which often denatures the protein of interest being purified. However, it has been shown that a short incubation in water above 70 °C will reversibly break the interaction (at least for biotinylated DNA) without denaturing streptavidin, allowing re-use of the streptavidin solid support.[7] A further application of streptavidin is for purification and detection of proteins genetically modified with the Strep-tag peptide. Streptavidin is widely used in Western blotting and immunoassays conjugated to some reporter molecule, such as horseradish peroxidase. Streptavidin has also been used in the developing field of Nanobiotechnology, the use of biological molecules such as proteins or lipids to create nanoscale devices/structures. In this context streptavidin can be used as a building block to link biotinylated DNA molecules to create single walled carbon nanotube scaffolds[8] or even complex DNA polyhedra.[9] The tetrameric streptavidin has also been used as a hub around which other proteins may be arranged, either by an affinity tag such as Strep-tag or AviTag or by genetic fusion to SpyTag.[10] Fusion to SpyTag allowed generation of assemblies with 8 or 20 streptavidin subunits. As well as a molecular force probe for atomic force microscopy studies,[11] novel materials such as 3D crystalline lattices[12] have also been created. Streptavidin has a mildly acidic isoelectric point (pI) of ~5, but a recombinant form of streptavidin with a near-neutral pI is also commercially available.
- Pretargeted Immunotherapy
Pretargeted immunotherapy uses streptavidin conjugated to a monoclonal antibody against cancer cell-specific antigens followed by an injection of radiolabelled biotin to deliver the radiation only to the cancerous cell. Initial hurdles involve saturation of the biotin binding sites on streptavidin with endogenous biotin instead of the injected radiolabelled biotin, and a high degree of radioactive exposure in the kidneys due to streptavidin’s strong cell adsorptive properties. It is currently thought that this high level of binding to adherent cell types, such as activated platelets and melanomas, is a result of integrin binding mediated through the RYD sequence in streptavidin.[13]
Variants with a controlled number of binding sites
- Monovalent vs. monomeric
Streptavidin is a tetramer and each subunit binds biotin with equal affinity. Multivalency is an advantage in some applications, for example where avidity effects improve the ability of molecules attached to streptavidin to detect specific T cells.[14] In other cases, such as the use of streptavidin for imaging specific proteins on cells, multivalency can perturb the function of the protein of interest. Monovalent streptavidin is an engineered recombinant form of streptavidin which is a tetramer but only one of the four binding sites is functional. This single binding site has 10−14 mol/L affinity and cannot cause cross-linking.[15] Applications of monovalent streptavidin have included fluorescent tracking of cell surface receptors, decorating DNA origami, and acting as a pointer to identify specific regions for cryo-electron microscopy.
Monomeric streptavidin is a recombinant form of streptavidin with mutations to break the tetramer into a monomer and to enhance the solubility of the resultant isolated subunit. Monomeric streptavidin versions have an affinity for biotin of 10−7mol/L 10−8mol/L and so are not ideal for labeling applications but are useful for purification, where reversibility is desirable.[16][17]
- Divalent
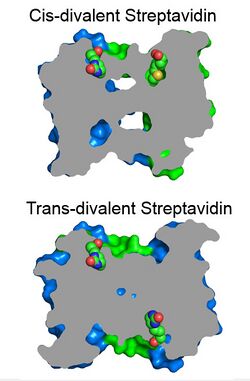
A streptavidin with exactly two biotin binding sites per tetramer can be produced by mixing subunits with and without a functional biotin binding site and purification by ion-exchange chromatography. The functional binding sites here have the same biotin binding stability as wild-type streptavidin. Divalent streptavidin with the two biotin binding sites together (cis-divalent) or apart (trans-divalent) can be separately purified.[18]
Comparison to avidin
Streptavidin is not the only protein capable of binding to biotin with high affinity. Avidin is the other most notable biotin-binding protein. Originally isolated from egg yolk, avidin only has 30% sequence identity to streptavidin, but almost identical secondary, tertiary and quaternary structure. It has a higher affinity for biotin (Kd ~ 10−15M) but in contrast to streptavidin, it is glycosylated, positively charged, has pseudo-catalytic activity (it can enhance the alkaline hydrolysis of an ester linkage between biotin and a nitrophenyl group) and has a higher tendency for aggregation. Also, streptavidin is the better biotin-conjugate binder; avidin has a lower binding affinity than streptavidin when biotin is conjugated to another molecule, despite avidin having the higher affinity for free, unconjugated biotin. Because streptavidin lacks any carbohydrate modification and has a near-neutral pI, it has the advantage of much lower nonspecific binding than avidin. Deglycosylated avidin (NeutrAvidin) is more comparable to the size, pI, and nonspecific binding of streptavidin.
See also
References
- ↑ "Avidin". Advances in Protein Chemistry 29: 85–133. 1975. doi:10.1016/s0065-3233(08)60411-8. PMID 237414.
- ↑ "Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation". Proceedings of the National Academy of Sciences of the United States of America 86 (7): 2190–4. April 1989. doi:10.1073/pnas.86.7.2190. PMID 2928324.
- ↑ "Structural origins of high-affinity biotin binding to streptavidin". Science 243 (4887): 85–8. January 1989. doi:10.1126/science.2911722. PMID 2911722.
- ↑ "The origins of femtomolar protein-ligand binding: hydrogen-bond cooperativity and desolvation energetics in the biotin-(strept)avidin binding site". Journal of the American Chemical Society 129 (17): 5419–29. May 2007. doi:10.1021/ja066950n. PMID 17417839.
- ↑ "A streptavidin variant with slower biotin dissociation and increased mechanostability". Nature Methods 7 (5): 391–3. May 2010. doi:10.1038/nmeth.1450. PMID 20383133.
- ↑ "Amine Landscaping to Maximize Protein-Dye Fluorescence and Ultrastable Protein-Ligand Interaction". Cell Chemical Biology 24 (8): 1040–1047.e4. August 2017. doi:10.1016/j.chembiol.2017.06.015. PMID 28757182.
- ↑ "The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures". Electrophoresis 26 (3): 501–10. February 2005. doi:10.1002/elps.200410070. PMID 15690449.
- ↑ "Biomolecule-directed assembly of self-supported, nanoporous, conductive, and luminescent single-walled carbon nanotube scaffolds". Small 8 (12): 1840–5. June 2012. doi:10.1002/smll.201102536. PMID 22461319.
- ↑ "DNA-directed three-dimensional protein organization". Angewandte Chemie 51 (14): 3382–5. April 2012. doi:10.1002/anie.201108710. PMID 22374892.
- ↑ "SpyAvidin hubs enable precise and ultrastable orthogonal nanoassembly". Journal of the American Chemical Society 136 (35): 12355–63. September 2014. doi:10.1021/ja505584f. PMID 25111182.
- ↑ "A nanoscale force probe for gauging intermolecular interactions". Angewandte Chemie 51 (8): 1903–6. February 2012. doi:10.1002/anie.201107210. PMID 22253141.
- ↑ "Generation of protein lattices by fusing proteins with matching rotational symmetry". Nature Nanotechnology 6 (9): 558–62. July 2011. doi:10.1038/nnano.2011.122. PMID 21804552.
- ↑ "Cell-adhesive properties of streptavidin are mediated by the exposure of an RGD-like RYD site". European Journal of Cell Biology 58 (2): 271–9. August 1992. PMID 1425765.
- ↑ "MHC/peptide tetramer-based studies of T cell function". Journal of Immunological Methods 268 (1): 21–8. October 2002. doi:10.1016/S0022-1759(02)00196-5. PMID 12213339.
- ↑ "A monovalent streptavidin with a single femtomolar biotin binding site". Nature Methods 3 (4): 267–73. April 2006. doi:10.1038/nmeth861. PMID 16554831.
- ↑ "Engineering soluble monomeric streptavidin with reversible biotin binding capability". The Journal of Biological Chemistry 280 (24): 23225–31. June 2005. doi:10.1074/jbc.M501733200. PMID 15840576.
- ↑ "Engineered streptavidin monomer and dimer with improved stability and function". Biochemistry 50 (40): 8682–91. October 2011. doi:10.1021/bi2010366. PMID 21892837.
- ↑ "Plug-and-play pairing via defined divalent streptavidins". Journal of Molecular Biology 426 (1): 199–214. January 2014. doi:10.1016/j.jmb.2013.09.016. PMID 24056174.
Further reading
- "Protein recognition of immobilized ligands: promotion of selective adsorption". Clinical Chemistry 33 (9): 1502–8. September 1987. PMID 3621554. http://www.clinchem.org/cgi/content/abstract/33/9/1502.
- Chodosh, Lewis A.; Buratowski, Stephen (2001). "Purification of DNA-Binding Proteins Using Biotin/Streptavidin Affinity Systems". Current Protocols in Protein Science. 9.7.1–9.7.13. doi:10.1002/0471140864.ps0907s12. ISBN 978-0-471-14086-3.
- "DNA stretching on functionalized gold surfaces". Nucleic Acids Research 22 (3): 492–7. February 1994. doi:10.1093/nar/22.3.492. PMID 8127690.
External links
- Swiss-Prot entry for Streptavidin precursor from Streptomyces avidinii
- Streptavidin at the US National Library of Medicine Medical Subject Headings (MeSH)
- Egg-stremely useful interaction QUite Interesting PDB Structure article at PDBe
Groups investigating and developing streptavidin or avidin-family proteins (Alphabetical order)
- Howarth lab (University of Oxford)
- Hytönen lab (University of Tampere)
- Kulomaa lab (University of Tampere)
- Livnah lab (Hebrew University of Jerusalem)
- Park Lab (University at Buffalo)
- Stenkamp lab (University of Washington)
- Wong lab (University of Calgary)