Biology:InterPro
 | |
|---|---|
| Content | |
| Description | InterPro functionally analyzes protein sequences and classifies them into protein families while predicting the presence of domains and functional sites. |
| Contact | |
| Research centre | EMBL |
| Laboratory | European Bioinformatics Institute |
| Primary citation | The InterPro protein families and domains database: 20 years on[1] |
| Release date | 1999 |
| Access | |
| Website | www |
| Download URL | ftp.ebi.ac.uk/pub/databases/interpro/ |
| Miscellaneous | |
| Data release frequency | 8-weekly |
| Version | 97.0 (9 November 2023) |
InterPro is a database of protein families, protein domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences[2] in order to functionally characterise them.[3][4]
The contents of InterPro consist of diagnostic signatures and the proteins that they significantly match. The signatures consist of models (simple types, such as regular expressions or more complex ones, such as Hidden Markov models) which describe protein families, domains or sites. Models are built from the amino acid sequences of known families or domains and they are subsequently used to search unknown sequences (such as those arising from novel genome sequencing) in order to classify them. Each of the member databases of InterPro contributes towards a different niche, from very high-level, structure-based classifications (SUPERFAMILY and CATH-Gene3D) through to quite specific sub-family classifications (PRINTS and PANTHER).
InterPro's intention is to provide a one-stop-shop for protein classification, where all the signatures produced by the different member databases are placed into entries within the InterPro database. Signatures which represent equivalent domains, sites or families are put into the same entry and entries can also be related to one another. Additional information such as a description, consistent names and Gene Ontology (GO) terms are associated with each entry, where possible.
Data contained in InterPro
InterPro contains three main entities: proteins, signatures (also referred to as "methods" or "models") and entries. The proteins in UniProtKB are also the central protein entities in InterPro. Information regarding which signatures significantly match these proteins are calculated as the sequences are released by UniProtKB and these results are made available to the public (see below). The matches of signatures to proteins are what determine how signatures are integrated together into InterPro entries: comparative overlap of matched protein sets and the location of the signatures' matches on the sequences are used as indicators of relatedness. Only signatures deemed to be of sufficient quality are integrated into InterPro. As of version 81.0 (released 21 August 2020) InterPro entries annotated 73.9% of residues found in UniProtKB with another 9.2% annotated by signatures that are pending integration.[5]
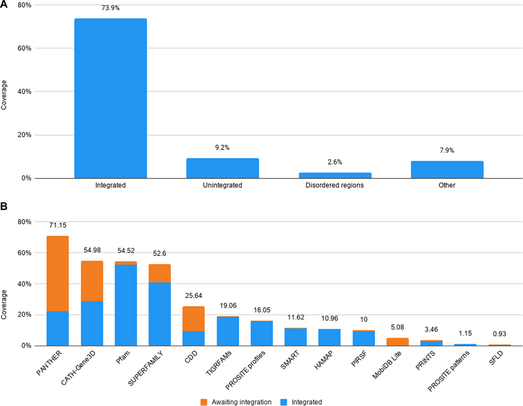
InterPro also includes data for splice variants and the proteins contained in the UniParc and UniMES databases.
InterPro consortium member databases
The signatures from InterPro come from 13 "member databases", which are listed below.
- CATH-Gene3D
- Describes protein families and domain architectures in complete genomes. Protein families are formed using a Markov clustering algorithm, followed by multi-linkage clustering according to sequence identity. Mapping of predicted structure and sequence domains is undertaken using hidden Markov models libraries representing CATH and Pfam domains. Functional annotation is provided to proteins from multiple resources. Functional prediction and analysis of domain architectures is available from the Gene3D website.
- CDD
- Conserved Domain Database is a protein annotation resource that consists of a collection of annotated multiple sequence alignment models for ancient domains and full-length proteins. These are available as position-specific score matrices (PSSMs) for fast identification of conserved domains in protein sequences via RPS-BLAST.
- HAMAP
- Stands for High-quality Automated and Manual Annotation of microbial Proteomes. HAMAP profiles are manually created by expert curators they identify proteins that are part of well-conserved bacterial, archaeal and plastid-encoded (i.e. chloroplasts, cyanelles, apicoplasts, non-photosynthetic plastids) proteins families or subfamilies.
- MobiDB
- MobiDB is database annotating intrinsic disorder in proteins.
- PANTHER
- PANTHER is a large collection of protein families that have been subdivided into functionally related subfamilies, using human expertise. These subfamilies model the divergence of specific functions within protein families, allowing more accurate association with function (human-curated molecular function and biological process classifications and pathway diagrams), as well as inference of amino acids important for functional specificity. Hidden Markov models (HMMs) are built for each family and subfamily for classifying additional protein sequences.
- Pfam
- Is large collection of multiple sequence alignments and hidden Markov models covering many common protein domains and families.
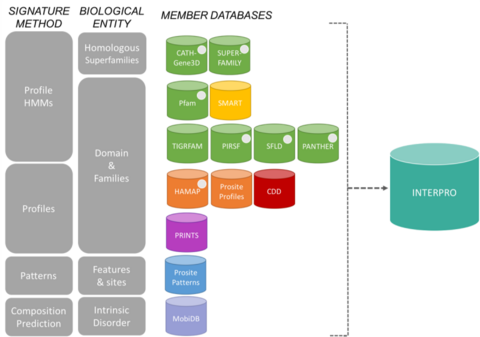
The 13 member databases of the InterPro consortium grouped by their signature construction method and the biological entity they focus on.[6] - PIRSF
- Protein classification system is a network with multiple levels of sequence diversity from superfamilies to subfamilies that reflects the evolutionary relationship of full-length proteins and domains. The primary PIRSF classification unit is the homeomorphic family, whose members are both homologous (evolved from a common ancestor) and homeomorphic (sharing full-length sequence similarity and a common domain architecture).
- PRINTS
- PRINTS is a compendium of protein fingerprints. A fingerprint is a group of conserved motifs used to characterise a protein family; its diagnostic power is refined by iterative scanning of UniProt. Usually the motifs do not overlap, but are separated along a sequence, though they may be contiguous in 3D-space. Fingerprints can encode protein folds and functionalities more flexibly and powerfully than can single motifs, their full diagnostic potency deriving from the mutual context afforded by motif neighbours.
- PROSITE
- PROSITE is a database of protein families and domains. It consists of biologically significant sites, patterns and profiles that help to reliably identify to which known protein family (if any) a new sequence belongs.
- SMART
- Simple Modular Architecture Research Tool Allows the identification and annotation of genetically mobile domains and the analysis of domain architectures. More than 800 domain families found in signaling, extracellular and chromatin-associated proteins are detectable. These domains are extensively annotated with respect to phyletic distributions, functional class, tertiary structures and functionally important residues.
- SUPERFAMILY
- SUPERFAMILY is a library of profile hidden Markov models that represent all proteins of known structure. The library is based on the SCOP classification of proteins: each model corresponds to a SCOP domain and aims to represent the entire SCOP superfamily that the domain belongs to. SUPERFAMILY has been used to carry out structural assignments to all completely sequenced genomes.
- SFLD
- A hierarchical classification of enzymes that relates specific sequence-structure features to specific chemical capabilities.
- TIGRFAMs
- TIGRFAMs is a collection of protein families, featuring curated multiple sequence alignments, hidden Markov models (HMMs) and annotation, which provides a tool for identifying functionally related proteins based on sequence homology. Those entries which are "equivalogs" group homologous proteins which are conserved with respect to function.
Data types
InterPro consists of seven types of data provided by different members of the consortium:
| Data Type | Description | Contributing Databases |
|---|---|---|
| InterPro Entries | Structural and/or functional domains of proteins predicted using one or more signatures | All 13 member databases |
| Member Database signatures | Signatures from member databases. These include signatures that are integrated into InterPro, and those that are not | All 13 member databases |
| Protein | Protein sequences | UniProtKB (Swiss-Prot and TrEMBL) |
| Proteome | Collection of proteins that belong to a single organism | UniProtKB |
| Structure | 3-dimensional structures of proteins | PDBe |
| Taxonomy | Protein taxonomic information | UniProtKB |
| Set | Groups of evolutionary related families | Pfam, CDD |

InterPro entry types
InterPro entries can be further broken down into five types:
- Homologous Superfamily: A group of proteins that share a common evolutionary origin as seen in their structural similarities, even if their sequences are not highly similar. These entries are specifically only provided by two member databases: CATH-Gene3D and SUPERFAMILY.
- Family: A group of proteins that have a common evolutionary origin determined through structural similarities, related functions, or sequence homology.
- Domain: A distinct unit in a protein with a particular function, structure, or sequence.
- Repeat: A sequence of amino acids, usually no longer than 50 amino acids, that tend to repeat many times in a protein.
- Site: A short sequence of amino acids where at least one amino acid is conserved. These include post-translation modification sites, conserved sites, binding sites, and active sites.
Access
The database is available for text- and sequence-based searches via a webserver, and for download via anonymous FTP. Like other EBI databases, it is in the public domain, since its content can be used "by any individual and for any purpose".[8] InterPro aims to release data to the public every 8 weeks, typically within a day of the UniProtKB release of the same proteins.
InterPro application programming interface (API)
InterPro provides an API for programmatic access to all InterPro entries and their related entries in Json format.[9] There are six main endpoints for the API corresponding to the different InterPro data types: entry, protein, structure, taxonomy, proteome and set.
InterProScan
InterProScan is a software package that allows users to scan sequences against member database signatures. Users can use this signature scanning software to functionally characterize novel nucleotide or protein sequences.[10] InterProScan is frequently used in genome projects in order to obtain a "first-pass" characterisation of the genome of interest.[11][12] As of December 2020[update], the public version of InterProScan (v5.x) uses a Java-based architecture.[13] The software package is currently only supported on a 64-bit Linux operating system.
InterProScan, along with many other EMBL-EBI bioinformatics tools, can also be accessed programmatically using RESTful and SOAP Web Services APIs.[14]
See also
References
- ↑ "The InterPro protein families and domains database: 20 years on". Nucleic Acids Research 49 (D1): D344–D354. November 2020. doi:10.1093/nar/gkaa977. PMID 33156333.
- ↑ "InterPro in 2011: new developments in the family and domain prediction database". Nucleic Acids Research 40 (Database issue): D306-12. January 2012. doi:10.1093/nar/gkr948. PMID 22096229.
- ↑ "The InterPro database, an integrated documentation resource for protein families, domains and functional sites". Nucleic Acids Research 29 (1): 37–40. January 2001. doi:10.1093/nar/29.1.37. PMID 11125043.
- ↑ "InterPro--an integrated documentation resource for protein families, domains and functional sites". Bioinformatics 16 (12): 1145–50. December 2000. doi:10.1093/bioinformatics/16.12.1145. PMID 11159333.
- ↑ 5.0 5.1 Blum, Matthias; Chang, Hsin-Yu; Chuguransky, Sara; Grego, Tiago; Kandasaamy, Swaathi; Mitchell, Alex; Nuka, Gift; Paysan-Lafosse, Typhaine et al. (2020-11-06). "The InterPro protein families and domains database: 20 years on" (in en). Nucleic Acids Research 49 (D1): D344–D354. doi:10.1093/nar/gkaa977. ISSN 0305-1048. PMID 33156333.
- ↑ EMBL-EBI. "Where does the data come from? | InterPro" (in en). https://www.ebi.ac.uk/training-beta/online/courses/interpro-functional-and-structural-analysis/what-is-interpro/where-does-data-come-from/.
- ↑ EMBL-EBI. "InterPro entry types | InterPro" (in en). https://www.ebi.ac.uk/training-beta/online/courses/interpro-functional-and-structural-analysis/what-is-an-interpro-entry/interpro-entry-types/.
- ↑ "Terms of Use for EMBL-EBI Services | European Bioinformatics Institute". http://www.ebi.ac.uk/Information/termsofuse.html.
- ↑ "How to download InterPro data? — InterPro Documentation". https://interpro-documentation.readthedocs.io/en/latest/download.html#interpro-application-programming-interface-api.
- ↑ "InterProScan: protein domains identifier" (Free full text). Nucleic Acids Research 33 (Web Server issue): W116-20. July 2005. doi:10.1093/nar/gki442. PMID 15980438.
- ↑ "Initial sequencing and analysis of the human genome". Nature 409 (6822): 860–921. February 2001. doi:10.1038/35057062. PMID 11237011. Bibcode: 2001Natur.409..860L. https://deepblue.lib.umich.edu/bitstream/2027.42/62798/1/409860a0.pdf.
- ↑ "The genome sequence of the malaria mosquito Anopheles gambiae". Science 298 (5591): 129–49. October 2002. doi:10.1126/science.1076181. PMID 12364791. Bibcode: 2002Sci...298..129H.
- ↑ "InterProScan 5: genome-scale protein function classification". Bioinformatics 30 (9): 1236–40. May 2014. doi:10.1093/bioinformatics/btu031. PMID 24451626.
- ↑ "The EMBL-EBI search and sequence analysis tools APIs in 2019". Nucleic Acids Research 47 (W1): W636–W641. July 2019. doi:10.1093/nar/gkz268. PMID 30976793.
External links
- — webserver
 |
