Biology:Interference (genetic)
Crossover interference is the term used to refer to the non-random placement of crossovers with respect to each other during meiosis. The term is attributed to Muller, who observed that one crossover "interferes with the coincident occurrence of another crossing over in the same pair of chromosomes, and I have accordingly termed this phenomenon ‘interference’."[1]
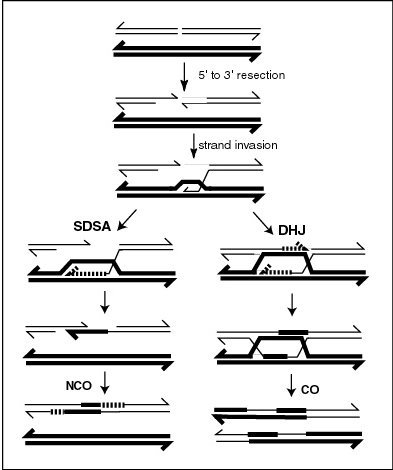
Meiotic crossovers (COs) appear to be regulated to ensure that COs on the same chromosome are distributed far apart (crossover interference). In the nematode worm Caenorhabditis elegans, meiotic double-strand breaks (DSBs) outnumber COs. Thus not all DSBs are repaired by a recombination process(es) leading to COs. The RTEL-1 protein is required to prevent excess meiotic COs. In rtel-1 mutants meiotic CO recombination is significantly increased and crossover interference appears to be absent.[2] RTEL1 likely acts by promoting synthesis-dependent strand annealing which results in non-crossover (NCO) recombinants instead of COs (see diagram).[2] Normally, about half of all DSBs are converted into NCOs. RTEL-1 appears to enforce meiotic crossover interference by directing the repair of some DSBs towards NCOs rather than COs.[2]
In humans, recombination rate increases with maternal age.[3] Furthermore, placement of female recombination events appears to become increasingly deregulated with maternal age, with a larger fraction of events occurring within closer proximity to each other than would be expected under simple models of crossover interference.[4]
High negative interference
Bacteriophage T4
High negative interference (HNI), in contrast to positive interference, refers to the association of recombination events ordinarily measured over short genomic distances, usually within a gene. Over such short distances there is a positive correlation (negative interference) of recombinational events. As studied in bacteriophage T4 this correlation is greater the shorter the interval between the sites used for detection.[5] HNI is due to multiple exchanges within a short region of the genome during an individual mating event.[6] What is counted as a “single exchange” in a genetic cross involving only distant markers may in reality be a complex event that is distributed over a finite region of the genome.[7] Switching between template DNA strands during DNA synthesis (see Figure, SDSA pathway), referred to as copy-choice recombination, was proposed to explain the positive correlation of recombination events within the gene.[8] HNI appears to require fairly precise base complementarity in the regions of the parental genomes where the associated recombination events occur.[9]
HIV
Each human immunodeficiency virus (HIV) particle contains two single-stranded positive sense RNA genomes. After infection of a host cell, a DNA copy of the genome is formed by reverse transcription of the RNA genomes. Reverse transcription is accompanied by template switching between the two RNA genome copies (copy-choice recombination).[10] From 5 to 14 recombination events per genome occur at each replication cycle.[11] This recombination exhibits HNI.[12] HNI is apparently caused by correlated template switches during minus-strand DNA synthesis.[13] Template switching recombination appears to be necessary for maintaining genome integrity and as a repair mechanism for salvaging damaged genomes.[10][14]
References
- ↑ Muller, H.J. (1916). "The mechanism of crossing over". Am. Nat. 50.
- ↑ 2.0 2.1 2.2 "RTEL-1 enforces meiotic crossover interference and homeostasis". Science 327 (5970): 1254–8. 2010. doi:10.1126/science.1183112. PMID 20203049.
- ↑ "Recombination rate and reproductive success in humans". Nat. Genet. 36 (11): 1203–6. 2004. doi:10.1038/ng1445. PMID 15467721.
- ↑ "Escape from crossover interference increases with maternal age". Nat Commun 6: 6260. 2015. doi:10.1038/ncomms7260. PMID 25695863.
- ↑ Chase M, Doermann AH. High Negative Interference over Short Segments of the Genetic Structure of Bacteriophage T4. Genetics. 1958 May;43(3):332-53. PMID: 17247760
- ↑ Edgar RS, Steinberg CM. On the origin of high negative interference over short segments of the genetic structure of bacteriophage T4. Virology. 1958 Aug;6(1):115-28. PMID: 13626191
- ↑ Steinberg CM, Edgar RS.A critical test of a current theory of genetic recombination in bacteriophage. Genetics. 1962 Feb;47:187-208. PMID: 13916671 PMCID:PMC1210322
- ↑ Bernstein H. On the mechanism of intragenic recombination. I. The rII region of bacteriophage T4. (1962) Journal of Theoretical Biology. 1962; 3, 335-353. https://doi.org/10.1016/S0022-5193(62)80030-7
- ↑ Berger H, Warren AJ. Effects of deletion mutations on high negative interference in T4D bacteriophage. Genetics. 1969 Sep;63(1):1-5. PMID:5365292 PMCID:PMC1212323
- ↑ 10.0 10.1 Rawson JMO, Nikolaitchik OA, Keele BF, Pathak VK, Hu WS. Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity. Nucleic Acids Res. 2018 Nov 16;46(20):10535-10545. doi: 10.1093/nar/gky910. PMID:30307534
- ↑ Cromer D, Grimm AJ, Schlub TE, Mak J, Davenport MP. Estimating the in-vivo HIV template switching and recombination rate. AIDS. 2016 Jan;30(2):185-92. Doi: 10.1097/QAD.0000000000000936. PMID:26691546
- ↑ Hu WS, Bowman EH, Delviks KA, Pathak VK. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J Virol. 1997 Aug;71(8):6028-36. PMID:9223494
- ↑ Anderson JA, Teufel RJ 2nd, Yin PD, Hu WS. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J Virol. 1998 Feb;72(2):1186-94. PMID: 9445017
- ↑ Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227-33. PMID: 1700865
External links

