Biology:Slippery sequence
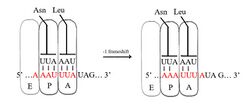
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame.[2][3][4][5][6] A −1 frameshift triggered by such a sequence is a Programmed −1 Ribosomal Frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins.[1]
The frameshift occurs due to wobble pairing. The Gibbs free energy of secondary structures downstream give a hint at how often frameshift happens.[7] Tension on the mRNA molecule also plays a role.[8] A list of slippery sequences found in animal viruses is available from Huang et al.[9]
Slippery sequences that cause a 2-base slip (−2 frameshift) have been constructed out of the HIV UUUUUUA sequence.[8]
See also
- Nucleic acid tertiary structure
- Open reading frame
- Ribosomal frameshifting
- Translational frameshift
- Transposable element
References
- ↑ 1.0 1.1 "Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region". Cell 55 (3): 447–58. November 1988. doi:10.1016/0092-8674(88)90031-1. PMID 2846182.
- ↑ "Characterization of the mechanical unfolding of RNA pseudoknots". Journal of Molecular Biology 375 (2): 511–28. January 2008. doi:10.1016/j.jmb.2007.05.058. PMID 18021801.
- ↑ "Stimulation of ribosomal frameshifting by antisense LNA". Nucleic Acids Research 38 (22): 8277–83. December 2010. doi:10.1093/nar/gkq650. PMID 20693527.
- ↑ "Dr Ian Brierley Research description". Department of Pathology, University of Cambridge. http://www.path.cam.ac.uk/research/investigators/brierley/research.html.
- ↑ "Molecular Biology: Frameshifting occurs at slippery sequences". Molecularstudy.blogspot.com. 2012-10-16. http://molecularstudy.blogspot.com/2012/10/frameshifting-occurs-at-slippery.html.
- ↑ "How translational accuracy influences reading frame maintenance". The EMBO Journal 18 (6): 1427–34. March 1999. doi:10.1093/emboj/18.6.1427. PMID 10075915.
- ↑ "Predicting ribosomal frameshifting efficiency". Physical Biology 5 (1): 016002. March 2008. doi:10.1088/1478-3975/5/1/016002. PMID 18367782. Bibcode: 2008PhBio...5a6002C.
- ↑ 8.0 8.1 "Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting". Nucleic Acids Research 40 (17): 8674–89. September 2012. doi:10.1093/nar/gks629. PMID 22743270.
- ↑ "A genome-wide analysis of RNA pseudoknots that stimulate efficient -1 ribosomal frameshifting or readthrough in animal viruses". BioMed Research International 2013: 984028. 2013. doi:10.1155/2013/984028. PMID 24298557.
External links
- Pseudobase
- Recode
- Frameshifting,+Ribosomal at the US National Library of Medicine Medical Subject Headings (MeSH)
- Wise2 - aligns a protein against a DNA sequence allowing frameshifts and introns
- FastY - compare a DNA sequence to a protein sequence database, allowing gaps and frameshifts
- Path - tool that compares two frameshift proteins (back-translation principle)
- Recode2 - Database of recoded genes, including those that require programmed Translational frameshift.
- Page for Coronavirus frameshifting stimulation element at Rfam
 |

