Biology:FAM210B
 Generic protein structure example |
FAM210B is a gene that which in Homo sapiens encodes the protein FAM210B. It has been conserved throughout evolutionary history, and is highly expressed in multiple tissues within the human body. FAM210B's primary location is the endoplasmic reticulum.
Gene
Locus
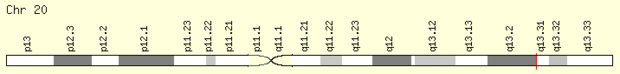
FAM210B is found on the plus strand of chromosome 20, precisely at 20q13.2. This gene is 9,749 bases in length, corresponding to a 192 amino acid protein.[1]
Aliases
Alternative names include C20orf108, Hypothetical protein LOC116151, DJ116H4.1, and 5A3.[1]
mRNA
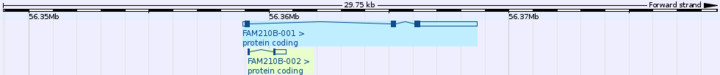
FAM210B has 2 known splice variants, formally known as FAM210B-001 an FAM210B-002. FAM210B-001 is 3 exons in length, whereas FAM210B-002 is 2 exons.[2]
Protein
The longest precursor protein is 192 amino acids in length. The molecular weight is 20.4 kdal, and isoelectric point is 10.8.[3]
Structure
Primary Structure
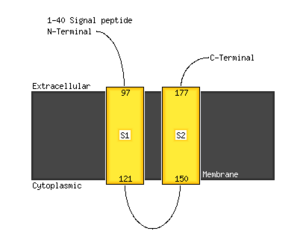
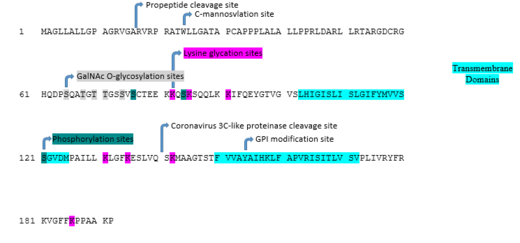
There is 1 positive charge run of 4 residues at position 79, EEKK. A high scoring transmembrane sequence is present at positions 99–123, VGVSLHIGISLISLGIFYM VVSSGV. An endoplasmic reticulum retention signal (PAAK) is present at positions 187–191.[3]
Secondary Structure
The majority of the structure is composed of alpha helices. As predicted my Phyre 2, 61% of the protein forms alpha helices, and 28% forms transmembrane segments. A detailed map of the transmembrane topology is shown below.[4]
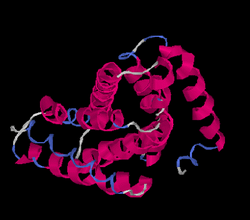
Tertiary Structure
I-Tasser predicted the folding of FAM210B to be as shown below.
Subcellular Location
The primary location of the FAM210B protein is the endoplasmic reticulum. The amino acid sequence contains the endoplasmic retention signal PAAK near the C-terminus. There are 2 transmembrane domains, which have been determined and reaffirmed by multiple methods. They are highlighted in the post-translational modification image below.[5]
Post-Translational Modifications
The following conceptual translation presents expected post-translational modifications. The modifications were predicted by MotifScan, and then searched in greater detail in Expasy[6] Modifications found include propeptide cleavage sites, C-mannosylation sites, GalNAc O-glycosylation sites, lysine glycation sites, phosphorylation sites, a Coronavirus 3C-like proteinase cleavage site, and a GPI modification site.
Homology
An important paralog for FAM210B is FAM210A. Orthologs and distant homologs have been found throughout mammalia, reptiles, birds, invertebrates, fish, amphibian, trichoplax, and fungi.[7]
Paralogs
| Gene Name | Common name | Accession number | Sequence length | E-value | Identity | Query cover |
| RCE1 protein | RCE1 | AAH52622.1 | 329 | 0.063 | 42% | 28% |
| Serine/Threonine-protein kinase | SBK2 | NP_001094871.2 | 348 | 1.3 | 54% | 15% |
| FAM210A | FAM210A | NP_689565.2 | 272 | 1.4 | 31% | 53% |
Orthologs
| Genus and species | Common name | Date of divergence (MYA) | Accession number | Sequence length | E-value | Identity | Query cover | Notes |
| Homo sapiens | Human | - | NP_543011.2 | 192 | 2.00E-138 | 100% | 100% | Mammal |
| Tarsius syrichta | Philippine tarsier | 67.6 | XP_008058424.1 | 150 | 3.00E-88 | 88% | 76% | Mammal |
| Fukomys damarensis | Damaraland mole-rat | 90.9 | XP_010627139.1 | 184 | 1.00E-77 | 82% | 76% | Mammal |
| Orninus orca | Killer whale | 97.5 | XP_004282340.1 | 192 | 2.00E-98 | 81% | 100% | Mammal |
| Camelus ferus | Bactrian camel | 97.5 | EPY80008.1 | 251 | 7.00E-72 | 87% | 69% | Mammal |
| Anolis carolinensis | Carolina anole | 320.5 | XP_003223790.1 | 215 | 4.00E-60 | 65% | 78% | Reptile |
| Thamnophis sirtalis | Common garter snake | 320.5 | XP_013919769.1 | 286 | 3.00E-59 | 67% | 71% | Reptile |
| Ophiophagus hannah | King cobra | 320.5 | ETE65490.1 | 152 | 8.00E-59 | 68% | 67% | Reptile |
| Cariama cristata | Reg-legged seriema | 320.5 | KRP59646.1 | 115 | 7.00E-58 | 78% | 59% | Bird |
| Egretta garzetta | Little egret | 320.5 | KFP21403.1 | 115 | 1.00E-57 | 77% | 59% | Bird |
| Anas platyrhynchos | Mallard | 320.5 | EOB08268.1 | 97 | 2.00E-14 | 44% | 50% | Bird |
| Gekko japonicus | Gekko | 320.5 | XP_015277771.1 | 304 | 1.00E-11 | 34% | 57% | Reptile |
| Xenopus tropicalis | Western clawed frog | 355.7 | NP_001072818.1 | 196 | 5.00E-57 | 55% | 94% | Amphibian |
| Xenopus laevis | African clawed-frog | 355.7 | NP_001088884.1 | 275 | 8.00E-10 | 29% | 53% | Amphibian |
| Callorhinchus milii | Australian ghostshark | 482.9 | XP_007910267.1 | 217 | 1.00E-48 | 73% | 55% | Fish |
| Drosophila sechellia | Fruit fly | 847 | XP_002037687.1 | 135 | 5.00E-31 | 48% | 71% | Invertebrate |
| Cerapachys biroi | Clonal raider ant | 847 | XP_011331931.1 | 344 | 3.00E-15 | 36% | 56% | Invertebrate |
| Ogataea parapolymorpha | Microorganism | 1302.5 | XP_013935759.1 | 208 | 7.00E-05 | 29% | 60% | Fungus |
| Populus trichocarpa | Black cottonwood | 1513.9 | XP_006379359.1 | 242 | 5.00E-04 | 33% | 49% | Plant |
| Trichoplax adhaerens | Trichoplax | Unknown | XP_002116161.1 | 149 | 5.00E-29 | 40% | 63% | Trichoplax |
| Mitosporidium daphinae | Parasite | Unknown | XP_013239484.1 | 162 | 1.00E-07 | 35% | 48% | Fungus |
Evolutionary History
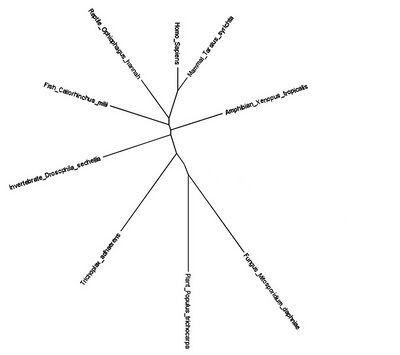
The size of the gene family is two, and includes FAM210A and FAM210B. Gene duplication seems to have occurred when plants diverged from protists. The most distant ortholog found was Populus trichocarpa, also known as the black cottonwood tree.[8] The unrooted phylogenetic tree below demonstrates this divergence.[9] All of the organisms in the tree can also be found in the ortholog table above.
Expression
FAM210B is ubiquitously expressed in Homo sapiens.[10] It is expressed in each stage of development, and in many compromised health states including breast, cervical, liver, lung, ovarian, and pancreatic tumors.[11]
Allen Brain Atlas
These images were taken from Allen Brain Atlas and demonstrate expression of FAM210B in the mouse brain. The images compare the expression of FAM210B (left) to dopamine beta hydroxylase (right) in a sagittal cut of the cerebellum.[12]
GEO Profiles
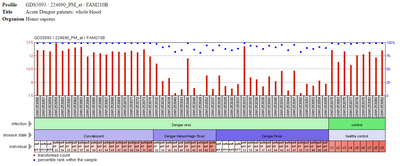
The following image contains the gene expression of FAM210B in patients with dengue fever or those who are convalescent. As seen by the blue dots, there is decreased rank of FAM210B in patients experiencing hemorrhagic fever and fever as compared to the healthy controls.[13]
The following image contains the gene expression of FAM210B in patients with severe bacterial pneumonia, severe influenza, and those who received the influenza vaccine. As seen in the image, FAM210B is lower in rank in the patients with severe bacterial pneumonia and are even lower in those with severe influenza, as compared to those who received the influenza vaccine.[14]
Interacting Proteins
The following proteins were determined to interact with FAM210B. Their interaction was determined by affinity capture and mass spectrometric methods.[15] By analyzing the function of these proteins, more insight can be given into the function of FAM210B.
| Protein | Function |
| HTR2C | Encodes transmembrane G-protein-coupled receptor. Responds to signaling through serotonin. |
| LPAR1 | Protein encoded by this gene is a LPA receptor. Mediate functions such as proliferation, platelet aggregation, smooth muscle contraction, tumor cell invasion. |
| REEP5 | May promote functional cell surface expression of olfactory receptors. |
| RTN1 | Reticulon encoding gene, associated with the ER and involved in neuroendocrine secretion. |
| RTN4 | Reticulon encoding gene, associated with the ER and involved in neuroendocrine secretion. |
| TM4SF20 | Interact with integrins to function cell adhesion, proliferation, and motility. |
| TSPAN5 | Mediate signal transduction events that regulate cell development, activation, growth, and motility. |
| TSPAN17 | Encodes member of transmembrane 4 superfamily, function is undetermined. |
| ATP6V0D1 | Encodes V-ATPase that mediates acidification of eukaryotic intracellular organelles. |
References
- ↑ 1.0 1.1 1.2 "FAM210B Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM210B.
- ↑ 2.0 2.1 "Gene: FAM210B (OTTHUMG00000032793) - Summary - Homo sapiens - Vega Genome Browser 64". http://vega.sanger.ac.uk/Homo_sapiens/Gene/Summary?db=core;g=OTTHUMG00000032793;r=20:56358915-56368663.
- ↑ 3.0 3.1 "SDSC Biology Workbench". http://workbench.sdsc.edu/.
- ↑ "Phyre 2 Results for FAM210B_Normal". http://www.sbg.bio.ic.ac.uk/phyre2/phyre2_output/d7f8c33c29cbb1de/summary.html.[yes|permanent dead link|dead link}}]
- ↑ "PSORT II Prediction". http://psort.hgc.jp/form2.html.
- ↑ "ExPASy: SIB Bioinformatics Resource Portal - Categories". https://www.expasy.org/proteomics/post-translational_modification.
- ↑ "BLAST: Basic Local Alignment Search Tool". http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "Nucleotide BLAST: Search nucleotide databases using a nucleotide query". http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome.
- ↑ "FigTree". http://tree.bio.ed.ac.uk/software/figtree/.
- ↑ "Family with sequence similarity 210, member B (FAM210B)". https://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?UGID=151287&TAXID=9606&SEARCH=FAM210B.
- ↑ "EST Profile - Hs.143736". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.143736.
- ↑ "Experiment Detail :: Allen Brain Atlas: Mouse Brain". http://mouse.brain-map.org/experiment/show/641524.
- ↑ "GDS5093 / 224693_PM_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS5093:224693_PM_at.
- ↑ "GDS3919 / ILMN_1708016". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS3919:ILMN_1708016.
- ↑ Lab, Mike Tyers. "FAM210B (PSEC0265) Result Summary | BioGRID". http://thebiogrid.org/125482/summary/homo-sapiens/fam210b.html.
 |