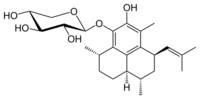
Chemistry:Pseudopterosin A
| |
| Names | |
|---|---|
| IUPAC name
(3S,7R,9S,9aR)-5-Hydroxy-3,6,9-trimethyl-7-(2-methylprop-1-en-1-yl)-2,3,7,8,9,9a-hexahydro-1H-phenalen-4-yl β-D-xylopyranoside
| |
| Systematic IUPAC name
(2S,3R,4S,5R)-2-{[(3S,7R,9S,9aR)-5-Hydroxy-3,6,9-trimethyl-7-(2-methylprop-1-en-1-yl)-2,3,7,8,9,9a-hexahydro-1H-phenalen-4-yl]oxy}oxane-3,4,5-triol | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEMBL | |
| ChemSpider | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C25H36O6 | |
| Molar mass | 432.557 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Pseudopterosin A is a diterpene glycoside isolated from the gorgonian sea whip Antillogorgia elisabethae, found in the Bahamas and Florida Keys.[1] Pseudopterosins A-D, which differ in the degree of acetylation at the sugar ring, were first isolated and reported in 1986.[2] There are at least 25 unique diterpenes isolated from this species of marine animal.[1] Samples of P. elisabethae from the Bahamas are found to have higher concentrations of pseudopterosins than populations from the Florida Keys, which have a greater diversity in diterpene structures.[3]
Uses
Pseudopterosins have anti-inflammatory and analgesic activity, with a mechanism of action different from the common non-steroidal anti-inflammatory drugs, NSAIDs.[4] Commercially, pseudopterosins are found in skin creams as topical anti-inflammatory agents.[5]
Biosynthesis
Elisabethatriene (2) has been identified as a key intermediate for the synthesis of the class of pseudopterosins and seco-pseudopterosins. A proposed mechanistic pathway for the synthesis of elisabethatriene from geranylgeranyl pyrophosphate (GGPP, 1), is described below. Elisabethatriene synthase, a diterpene cyclase enzyme, catalyzes the transformation of the diterpene GGPP to a 10-membered carbon skeleton followed by hydride migration towards the bicyclic ring system.[1][6] This cyclase enzyme has been identified as a key enzyme in forming the carbon skeleton of pseudopterosins in one step. An alternative mechanism has been proposed in which a six-membered ring is formed first, then a second ring closing for the bicyclic system.[6]
The biosynthesis of the pseudopterosins continues with an aromatization to erogorgiaene (3), two oxidations to a dihyroxyerogorgiaene (4, then 5), and another oxidation to an ortho-hydroxyquinone (6). Ring closure (7), re-aromatization to (8) and glycosylation yield Pseudopterosin A (9). This is a plausible biosynthetic pathway, and intermediates 2, 3, 6, 7, and 8 have been identified using radio labeling studies.[1] An alternative mechanism has been proposed with no hydroxyquinone intermediate (6). Rather, molecule 3 undergoes two subsequent oxidations at C-6 and C-7 to a structure resembling 8, then glycosylation to pseudopterosin.[6]
The branching point for the biosynthesis of the tricyclic pseudopterosins versus the bicyclic seco-pseudopterosins occurs at compound 11, the aromatized bicycle erogorgiaene. 11 is oxidized once then hydroxylated followed by glycosylation to make the bicyclic seco-pseudopterosins.
The proposed synthesis of artificial anti-inflammatory metabolites is modeled after pseudopterosins and is based on the bicyclic seco-pseudopterosin structure 6.[1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 A. Kohl, A. Ata, R. Kerr. J. Ind Microbiol Biotechnol (2003) 30: 495-499.
- ↑ S. Look, W. Fenical, G. Matsumoto and J. Clardy. J. Org. Chem. (1986) 51: 5140-5145
- ↑ A. Kohl and R. Kerr. Mar. Drugs (2003) 1: 54-65.
- ↑ A. Kohl, R. Kerr. Arch. of Biochem. and Biophys. (2004) 424: 97-104.
- ↑ A. Mayer, P. Jacobson, W. Fenical, R. Jabocs and K. Glaser. Life Sciences (1998) 62: 401-407.
- ↑ 6.0 6.1 6.2 R. Kerr, A. Kohl, and T. Ferns. J. Ind Microbiol Biotech (2006) 33: 532-538.
 |

