Chemistry:Crystalline sponges
Crystalline sponges are series of organometallic networks developed by Japanese chemist Makoto Fujita.[1][2] The organic small molecules are absorbed into the void space of the crystalline sponges. Since the organometallic network of crystalline sponges can interact with the small molecule substrates via non-covalent interactions, the absorption can be selective. That is, the crystalline sponge can enrich certain molecules from a mixture.[3] As the crystal sponges are highly organized frameworks, the structure of the whole host-guest complex can be characterized by X-ray diffraction. Because the absorbate is encapsulated in a pre-organized environment, no single crystal of the substrate is needed in the X-ray diffraction. Besides, the X-ray crystallography of liquid samples can also be conducted.[4]
Molecular structure and composition
The first crystalline sponges developed by Makoto Fujita is [(Co(NCS)2)3(TPT)4], which is an infinitely extensive framework of Co2+ octahedral complex. Each octahedral complex is composed of six Co(NCS) vertexes and four 2,4,6-tris(4-pyridyl)-1,3,5-triazine (TPT) ligands. Besides the basic octahedral cavity (M6L4), there are also two different cuboctahedral cavities (M12L8 and M12L24) which can accommodate larger molecules, such as C60, C70.[3]
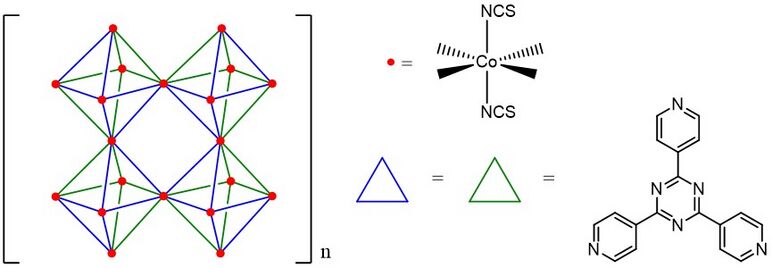
In 2013, the Fujita and his team discovered that the [(ZnI2)3(TPT)2] organometallic network can also act as a crystalline sponge.[4][5] Because Zn crystalline sponges are less symmetrical (C2) than Co crystalline sponges, the X-ray diffraction analysis of its guest molecules is easier to be elucidated.[6]
The Zn-based sponges also have following advantages: (1) Pores sizes are suitable for accommodating general organic compounds (5 × 8Å), (2) The distribution of TPT ligand is flat, providing better stacking opportunity of aromatic guest compounds or C—H π interaction opportunity with aliphatic guest compounds, (3) The iodine atoms and the pyridinium protons can be hydrogen bond acceptors or donors, respectively. They would enhance the interaction between the substrate and the crystalline sponges, (4) The framework of Zn-based crystalline sponge is flexible to some extent. The molecule which is a little bit larger than the sponge cavity can still be accommodated via framework expansion to adjust the pore size.[6]
Preparation
For preparation of [(Co(NCS)2)3(TPT)4] crystalline sponges, the methanol solution of Co(NCS)2 was added into the TPT solution in 1,2-dichlorobenzene/methanol. After 7 days, the [(Co(NCS)2)3(TPT)4] would form and can be isolated by filtration.
For preparation of [(ZnI2)3(TPT)2] crystalline sponges, the methanol solution of ZnI2 was added onto the top of TPT in nitrobenzene solution. After 7 days, the [(ZnI2)3(TPT)2] would form and can be isolated by filtration.[5][6]
Once the feedstocks were mixed, crystalline sponges would be produced via self-assembling processes, forming the thermodynamically stable organized network structures.
Application
Extract compounds from mixture
The crystalline sponges can be viewed as the extension of organometallic cage compounds. They can not only accommodate guest molecules, but also provide non-covalent interactions with the guest.[7] To exploit these properties, the Fujita group demonstrates [(Co(NCS)2)3(TPT)4] can enrich C70, C78, or other higher fullerenes from C60 mixtures as normal cage compounds did in precedent research.[3][8]
X-ray crystallography of liquids encapsulated within the sponge framework
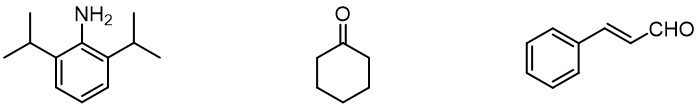
In 2013, the Fujita group rediscovered that the molecular structure of liquids can be elucidated by X-ray crystallography in the presence of [(ZnI2)3(TPT)2] crystalline sponges. The guest compounds, such as cyclohexanone, cinnamaldehyde were dropped onto the pre-made single crystal of crystalline sponges. The liquid samples would penetrate into the crystalline sponges, and occupy the sponges’ cavities. As crystalline sponges provide a well-organized structure, the guests in sponges’ cavities would also have organized distribution in the space. As a result, the structure of guest can be characterized by X-ray crystallography .[4]
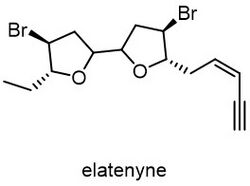
Some of the structures of liquid natural products which are difficult to differentiate by NMR spectroscopy can be easily characterized by crystalline sponges techniques. Elatenyne is a liquid molecule isolated from Laurencia elata, marine red algae.[9] Due to its pseudo-C2 symmetry, the difference of NMR spectrum between elatenyne and its stereoisomer is hard to differentiate.[10][11] However, with the crystalline sponges, the Fujita group easily elucidated the chirality in the elatenyne.[11]
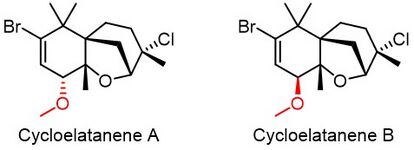
Cycloelatanene A and B are a pair of diastereomers isolated from Laurencia elata.[12] Both compounds are liquid, so the traditional X-ray diffraction analysis cannot elucidate their absolute chirality. After the crystalline sponges assisted X-ray crystallography, the Fujita group revised the chirality of C4 position reported by precedent NMR analysis.[12][13][14]
References
- ↑ Rosenberger, Lara; Essen, Carolina von; Khutia, Anupam; Kühn, Clemens; Urbahns, Klaus; Georgi, Katrin; Hartmann, Rolf W.; Badolo, Lassina (2020-07-01). "Crystalline Sponges as a Sensitive and Fast Method for Metabolite Identification: Application to Gemfibrozil and its Phase I and II Metabolites" (in en). Drug Metabolism and Disposition 48 (7): 587–593. doi:10.1124/dmd.120.091140. PMID 32434832. https://dmd.aspetjournals.org/content/48/7/587. Retrieved 2021-12-03.
- ↑ Inokuma, Yasuhide; Yoshioka, Shota; Ariyoshi, Junko; Arai, Tatsuhiko; Hitora, Yuki; Takada, Kentaro; Matsunaga, Shigeki; Rissanen, Kari et al. (2013-03-28). "X-ray analysis on the nanogram to microgram scale using porous complexes" (in en). Nature 495 (7442): 461–466. doi:10.1038/nature11990. ISSN 0028-0836. PMID 23538828. Bibcode: 2013Natur.495..461I. http://www.nature.com/articles/nature11990.
- ↑ 3.0 3.1 3.2 3.3 Inokuma, Y.; Arai, T.; Fujita, M. (2010). "Networked molecular cages as crystalline sponges for fullerenes and other guests". Nat. Chem. 2 (9): 780–783. doi:10.1038/nchem.742. PMID 20729900. Bibcode: 2010NatCh...2..780I. https://www.nature.com/articles/nchem.742?message-global=remove&hilite_compound=true.
- ↑ 4.0 4.1 4.2 Inokuma, Y.; Yoshioka, S.; Ariyoshi, J.; Arai, T.; Hitora, Y.; Takada, K.; Matsunaga, S.; Rissanen, K.; Fujita, M. (2013). "X-ray analysis on the nanogram to microgram scale using porous complexes". Nature 495 (7442): 461–466. doi:10.1038/nature11990. PMID 23538828. Bibcode: 2013Natur.495..461I. https://www.nature.com/articles/nature11990.
- ↑ 5.0 5.1 Biradha, K.; Fujita, M. (2002). "A Springlike 3D-Coordination Network That Shrinks or Swells in a Crystal-to-Crystal Manner upon Guest Removal or Readsorption". Angew. Chem. Int. Ed. 41 (18): 3392–3395. doi:10.1002/1521-3773(20020916)41:18<3392::AID-ANIE3392>3.0.CO;2-V. PMID 12298042. https://onlinelibrary.wiley.com/doi/10.1002/1521-3773(20020916)41:18%3C3392::AID-ANIE3392%3E3.0.CO;2-V.
- ↑ 6.0 6.1 6.2 Hoshino, M.; Khutia, A.; Xing, H.; Inokuma, Y.; Fujita, M. (2016). "The crystalline sponge method updated". IUCrJ 3 (Pt 2): 139–151. doi:10.1107/S2052252515024379. PMID 27006777. PMC 4775162. https://journals.iucr.org/m/issues/2016/02/00/de5035/index.html.
- ↑ Yoshizawa, M.; Klosterman, J. K.; Fujita, M. (2009). "Functional Molecular Flasks: New Properties and Reactions within Discrete, Self-Assembled Hosts". Angew. Chem. Int. Ed. 48 (19): 3418–3438. doi:10.1002/anie.200805340. PMID 19391140. https://onlinelibrary.wiley.com/doi/full/10.1002/anie.200805340.
- ↑ Huerta, E.; Metselaar, G. A.; Fragoso, A.; Santos, E.; Bo, C.; de Mendoza, J. (2007). "Selective Binding and Easy Separation of C70 by Nanoscale Self-Assembled Capsules". Angew. Chem. Int. Ed. 46 (1–2): 202–205. doi:10.1002/anie.200603223. PMID 17031895. https://onlinelibrary.wiley.com/doi/full/10.1002/anie.200603223.
- ↑ Hall, J. G.; Reiss, J. A. (1986). "Elatenyne—a Pyrano[3,2-BPyranyl Vinyl Acetylene From the Red Alga Laurencia elata"]. Aust. J. Chem. 39 (9): 1401–1409. doi:10.1071/ch9861401. https://www.publish.csiro.au/ch/CH9861401.
- ↑ Dyson, B. S.; Burton, J. W.; Sohn, T.-I.; Kim, B.; Bae, H.; Kim, D. (2012). "Total Synthesis and Structure Confirmation of Elatenyne: Success of Computational Methods for NMR Prediction with Highly Flexible Diastereomers". J. Am. Chem. Soc. 134 (28): 11781−11790. doi:10.1021/ja304554e. PMID 22758928. https://pubs.acs.org/doi/abs/10.1021/ja304554e.
- ↑ 11.0 11.1 Urban, S.; Brkljača, R.; Hoshino, M.; Lee, S.; Fujita, M. (2016). "Determination of the Absolute Configuration of the Pseudo-Symmetric Natural Product Elatenyne by the Crystalline Sponge Method". Angew. Chem. Int. Ed. 55 (8): 2678–2682. doi:10.1002/anie.201509761. PMID 26880368. https://onlinelibrary.wiley.com/doi/full/10.1002/anie.201509761.
- ↑ 12.0 12.1 Dias, D. A.; Urban, S. (2011). "Phytochemical studies of the southern Australian marine alga, Laurencia elata". Phytochemistry 72 (16): 2081–2089. doi:10.1016/j.phytochem.2011.06.012. PMID 21802699. Bibcode: 2011PChem..72.2081D. https://www.sciencedirect.com/science/article/pii/S0031942211003098.
- ↑ Lee, S.; Hoshino, M.; Fujita, M.; Urban, S. (2017). "Cycloelatanene A and B: absolute configuration determination and structural revision by the crystalline sponge method". Chem. Sci. 8 (2): 1547–1550. doi:10.1039/C6SC04288K. PMID 28572911.
- ↑ Inokuma, Y.; Yoshioka, S.; Ariyoshi, J.; Arai, T.; Hitora, Y.; Takada, K.; Matsunaga, S.; Rissanen, K. et al. (2013). "CCDC database". Nature 495 (7442): 461–466. doi:10.1038/nature11990. PMID 23538828. Bibcode: 2013Natur.495..461I. https://www.ccdc.cam.ac.uk/structures/Search?Doi=10.1038%2Fnature11990&DatabaseToSearch=Published.
 |