Biology:Inbreeding avoidance
Inbreeding avoidance, or the inbreeding avoidance hypothesis, is a concept in evolutionary biology that refers to the prevention of the deleterious effects of inbreeding. Animals only rarely exhibit inbreeding avoidance.[1] The inbreeding avoidance hypothesis posits that certain mechanisms develop within a species, or within a given population of a species, as a result of assortative mating and natural and sexual selection, in order to prevent breeding among related individuals. Although inbreeding may impose certain evolutionary costs, inbreeding avoidance, which limits the number of potential mates for a given individual, can inflict opportunity costs.[2] Therefore, a balance exists between inbreeding and inbreeding avoidance. This balance determines whether inbreeding mechanisms develop and the specific nature of such mechanisms.[3]
A 2007 study showed that inbred mice had significantly reduced survival when they were reintroduced into a natural habitat.[4]
Inbreeding can result in inbreeding depression, which is the reduction of fitness of a given population due to inbreeding. Inbreeding depression occurs via appearance of disadvantageous traits due to the pairing of deleterious recessive alleles in a mating pair's progeny.[5] When two related individuals mate, the probability of deleterious recessive alleles pairing in the resulting offspring is higher as compared to when non-related individuals mate because of increased homozygosity. However, inbreeding also gives opportunity for genetic purging of deleterious alleles that otherwise would continue to exist in population and could potentially increase in frequency over time. Another possible negative effect of inbreeding is weakened immune system due to less diverse immunity alleles as a result of outbreeding depression.[6]
A review of the genetics of inbreeding depression in wild animal and plant populations, as well as in humans, led to the conclusion that inbreeding depression and its opposite, heterosis (hybrid vigor), are predominantly caused by the presence of recessive deleterious alleles in populations.[7] Inbreeding, including self-fertilization in plants and automictic parthenogenesis (thelytoky) in hymenoptera, tends to lead to the harmful expression of deleterious recessive alleles (inbreeding depression). Cross-fertilization between unrelated individuals ordinarily leads to the masking of deleterious recessive alleles in progeny.[8][9]
Many studies have demonstrated that homozygous individuals are often disadvantaged with respect to heterozygous individuals.[10] For example, a study conducted on a population of South African cheetahs demonstrated that the lack of genetic variability among individuals in the population has resulted in negative consequences for individuals, such as a greater rate of juvenile mortality and spermatozoal abnormalities.[11] When heterozygotes possess a fitness advantage relative to a homozygote, a population with a large number of homozygotes will have a relatively reduced fitness, thus leading to inbreeding depression. Through these described mechanisms, the effects of inbreeding depression are often severe enough to cause the evolution of inbreeding avoidance mechanisms.[12]
Mechanisms
Inbreeding avoidance mechanisms have evolved in response to selection against inbred offspring. Inbreeding avoidance occurs in nature by at least four mechanisms: kin recognition, dispersal, extra-pair/extra-group copulations, and delayed maturation/reproductive suppression.[3][12] These mechanisms are not mutually exclusive and more than one can occur in a population at a given time.
Kin recognition

Kin recognition is the mechanism by which individuals identify and avoid mating with closely related conspecifics. There have been numerous documented examples of instances in which individuals are shown to find closely related conspecifics unattractive. In one set of studies, researchers formed artificial relative and non-relative mate-pairs (artificial meaning they preferentially paired individuals to mate for the purposes of the experiments) and compared the reproductive results of the two groups. In these studies, paired relatives demonstrated reduced reproduction and higher mating reluctance when compared with non-relatives.[12][13][14][15] For example, in a study by Simmons in field crickets, female crickets exhibited greater mating latency for paired siblings and half-siblings than with non-siblings.[13] In another set of studies, researchers allowed individuals to choose their mates from conspecifics that lie on a spectrum of relatedness. In this set, individuals were more likely to choose non-related over related conspecifics.[12][14][16] For example, in a study by Krackow et al., male wild house mice were set up in an arena with four separate openings leading to cages with bedding from conspecifics. The conspecifics exhibited a range of relatedness to the test subjects, and the males significantly preferred the bedding of non-siblings to the bedding of related females.[14]
Studies have shown that kin recognition is more developed in species in which dispersal patterns facilitate frequent adult kin encounters.[12]
There is a significant amount of variation in the mechanisms used for kin recognition. These mechanisms include recognition based on association or familiarity, an individual's own phenotypic cues, chemical cues, and the MHC genes. In association/familiarity mechanisms, individuals learn the phenotypic profiles of their kin and use this template for kin recognition.[12] Many species accomplish this by becoming "familiar" with their siblings, litter mates, or nestmates. These species rely on offspring being reared in close proximity to achieve kin recognition. This is called the Westermarck effect.[17] For example, Holmes and Sherman conducted a comparative study in Arctic ground squirrels and Belding's ground squirrels. They manipulated the reared groups to include both siblings and cross-fostered nestmates and found that in both species the individuals were equally aggressive toward their nestmates, regardless of kinship.[18] In certain species where social groups are highly stable, relatedness and association between infants and other individuals are usually highly correlated.[12][19] Therefore, degree of association can be used as a meter for kin recognition.
Individuals can also use their own characteristics or phenotype as a template in kin recognition. For example, in one study, Mateo and Johnston had golden hamsters reared with only non-kin then later had them differentiate between odors of related and non-related individuals without any postnatal encounters with kin. The hamsters were able to discriminate between the odors, demonstrating the use of their own phenotype for the purpose of kin recognition.[20] This study also provides an example of a species utilizing chemical cues for kin recognition.
The major histocompatibility complex genes, or MHC genes, have been implicated in kin recognition.[21] One idea is that the MHC genes code for a specific pheromone profile for each individual, which are used to discriminate between kin and non-kin conspecifics. Several studies have demonstrated the involvement of the MHC genes in kin recognition. For example, Manning et al. conducted a study in house mice that looked at the species's behavior of communal nesting, or nursing one's own pups as well as the pups of other individuals. As Manning et al. state, kin selection theory predicts that the house mice will selectively nurse the pups of their relatives in order to maximize inclusive fitness. Manning et al. demonstrate that the house mice utilize the MHC genes in the process of discriminating between kin by preferring individuals who share the same allelic forms the MHC genes.[22]
Human kin recognition
The possible use of olfaction-biased mechanisms in human kin recognition and inbreeding avoidance was examined in three different types of study.[23] The results indicated that olfaction may help mediate the development during childhood of incest avoidance (the Westermarck effect).
Post-copulatory inbreeding avoidance in mice
Experiments using in vitro fertilization in the mouse, provided evidence of sperm selection at the gametic level.[24] When sperm of sibling and non-sibling males were mixed, a fertilization bias towards the sperm of the non-sibling males was observed. The results were interpreted as egg-driven sperm selection against related sperm.
Inbreeding avoidance in plants
Experiments were performed with the dioecious plant Silene latifolia to test whether post-pollination selection favors less related pollen donors and reduces inbreeding.[25] The results showed that in S. latifolia, and presumably in other plant systems with inbreeding depression, pollen or embryo selection after multiple-donor pollination may reduce inbreeding.
Dispersal
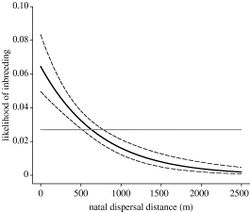
Some species will adopt dispersal as a way to separate close relatives and prevent inbreeding.[12] The initial dispersal route species may take is known as natal dispersal, whereby individuals move away from the area of birth. Subsequently, species may then resort to breeding dispersal, whereby individuals move from one non-natal group to another. Nelson-Flower et al. (2012) conducted a study on southern pied babblers and found that individuals may travel farther distances from natal groups than from non-natal groups.[26] This may be attributed to the possibility of encountering kin within local ranges when dispersing. The extent to which an individual in a particular species will disperse depends on whether the benefits of dispersing can outweigh both the costs of inbreeding and the costs of dispersal. Long‐distance movements can bear mortality risks and energetic costs.[27]
Sex-biased dispersal
In many cases of dispersal, one sex shows a greater tendency to disperse from their natal area than the opposite sex.[28] The extent of bias for a particular sex is dependent on numerous factors which include, but are not limited to: mating system, social organization, inbreeding and dispersal costs, and physiological factors.[27][28][29][30] When the costs and benefits of dispersal are symmetric for both males and females, then no sex-biased dispersal is expected to be observed in species.[27]
Female dispersal
Birds tend to adopt monogamous mating systems in which the males remain in their natal groups to defend familiar territories with high resource quality.[28] Females generally have high energy expenditure when producing offspring, therefore inbreeding is costly for the females in terms of offspring survival and reproductive success. Females will then benefit more by dispersing and choosing amongst these territorial males. In addition, according to the Oedipus hypothesis, daughters of female birds can cheat their mothers through brood parasitism, therefore females will evict the females from the nest, forcing their daughters to disperse. Female dispersal is not seen only in birds; males may remain philopatric in mammals when the average adult male residency in a breeding group exceeds the average age for female maturation and conception.[30] For example, in a community of chimpanzees in Gombe National Park, males tend to remain in their natal community for the duration of their lives, while females typically move to other communities as soon as they reach maturity.[31]
Male dispersal
Male dispersal is more common in mammals with cooperative breeding and polygynous systems. Australian marsupial juvenile males have a greater tendency to disperse from their natal groups, while the females remain philopatric.[32] In Antechinus this is due to the fact that males die immediately after mating; therefore when they disperse to mate, they often meet with female natal groups with zero males present. Furthermore, the Oedipus hypothesis also states that fathers in polygynous systems will evict sons with the potential to cuckold them.[28] Polygynous mating systems also influence intrasexual competition between males, where in cases where males can guard multiple females and exert their dominance, subordinate males are often forced to disperse to other non-natal groups.
When species adopt alternative inbreeding avoidance mechanisms, they can indirectly influence whether a species will disperse. Their choice for non-natal group males then selects for male dispersal.
Delayed maturation
The delayed sexual maturation of offspring in the presence of parents is another mechanism by which individuals avoid inbreeding. Delayed maturation scenarios can involve the removal of the original, opposite-sex parent, as is the case in female lions that exhibit estrus earlier following the replacement of their fathers with new males. Another form of delayed maturation involves parental presence that inhibits reproductive activity, such as in mature marmosets offspring that are reproductively suppressed in the presence of opposite sex parents and siblings in their social groups.[12] Reproductive suppression occurs when sexually mature individuals in a group are prevented from reproducing due to behavioral or chemical stimuli from other group members that suppress breeding behavior.[33] Social cues from the surrounding environment often dictate when reproductive activity is suppressed and involves interactions between same-sex adults. If the current conditions for reproduction are unfavorable, such as when presented with only inbreeding as a means to reproduce, individuals may increase their lifetime reproductive success by timing their reproductive attempts to occur during more favorable conditions. This can be achieved by individuals suppressing their reproductive activity in poor reproduction conditions.
Inbreeding avoidance between philopatric offspring and their parents/siblings severely restricts breeding opportunities of subordinates living in their social groups. A study by O'Riain et al. (2000) examined meerkats social groups and factors affecting reproductive suppression in subordinate females. They found that in family groups, the absence of a dominant individual of either sex led to reproductive quiescence. Reproductive activity only resumed upon another sexually mature female obtaining dominance, and immigration of an unrelated male. Reproduction required both the presence of an unrelated opposite-sex partner, which acted as appropriate stimulus on reproductively suppressed subordinates that were quiescent in the presence of the original dominant individual.[33]

Extra-pair copulations
In various species, females benefit by mating with multiple males, thus producing more offspring of higher genetic diversity and potentially quality. Females that are pair bonded to a male of poor genetic quality, as can be the case in inbreeding, are more likely to engage in extra-pair copulations in order to improve their reproductive success and the survivability of their offspring.[34] This improved quality in offspring is generated from either the intrinsic effects of good genes, or from interactions between compatible genes from the parents. In inbreeding, loss of heterozygosity contributes to the overall decreased reproductive success, but when individuals engage in extra-pair copulations, mating between genetically dissimilar individuals leads to increased heterozygosity.[35]
Extra-pair copulations involve a number of costs and benefits for both male and female animals. For males, extra-pair copulation involves spending more time away from the original pairing in search of other females. This risks the original female being fertilized by other males while the original male is searching for partners, leading to a loss of paternity. The tradeoff for this cost depends entirely on whether the male is able to fertilize the other females’ eggs in the extra-pair copulation. For females, extra-pair copulations ensure egg fertilization, and provide enhanced genetic variety with compatible sperm that avoid expression of damaging recessive genes that come with inbreeding.[36] Through extra-pair mating, females are able to maximize the genetic variability of their offspring, providing protection against environmental changes that may otherwise target more homozygous populations that inbreeding often produces.[37]
Whether a female engages in extra-pair copulations for the sake of inbreeding avoidance depends on whether the costs of extra-pair copulation outweigh the costs of inbreeding. In extra-pair copulations, both inbreeding costs and pair-bond male loss (leading to the loss of paternal care) must be considered with the benefits of reproductive success that extra-pair copulation provides. When paternal care is absent or has little influence on offspring survivability, it is generally favorable for females to engage in extra-pair mating to increase reproductive success and avoid inbreeding.[34]
Gaps
Inbreeding avoidance has been studied via three main methods: (1) observing individual behavior in the presence and absence of close kin, (2) contrasting costs of avoidance with costs of tolerating close inbreeding, and (3) comparing observed and random frequencies of close inbreeding.[38] No method is perfect, giving rise to questions about the completeness and consistency of the inbreeding avoidance hypothesis.[38][39] Although the first option, individual behavioral observation, is preferred and most widely used, there is still debate over whether it can provide definitive evidence for inbreeding avoidance.
A majority of the literature on inbreeding avoidance was published at least 15 years ago, allowing for growth and development of the study through contemporary experimental methods and technology. Molecular techniques such as DNA fingerprinting have become more advanced and accessible, improving the efficiency and accuracy of measuring relatedness.[12] Studying inbreeding avoidance in carnivores has garnered increased interest due to ongoing work to explain their social behaviors.[40]
References
- ↑ de Boer, Raïssa A.; Vega-Trejo, Regina; Kotrschal, Alexander; Fitzpatrick, John L. (July 2021). "Meta-analytic evidence that animals rarely avoid inbreeding" (in en). Nature Ecology & Evolution 5 (7): 949–964. doi:10.1038/s41559-021-01453-9. ISSN 2397-334X. PMID 33941905. https://www.nature.com/articles/s41559-021-01453-9.
- ↑ "When should animals tolerate inbreeding?". American Naturalist 128 (4): 529–537. 1986. doi:10.1086/284585.
- ↑ 3.0 3.1 "Behavioural inbreeding avoidance in wild African elephants". Molecular Ecology 16 (19): 4138–4148. 2007. doi:10.1111/j.1365-294x.2007.03483.x. PMID 17784925.
- ↑ "An experimental study of inbreeding depression in a natural habitat". Science 266 (5183): 271–273. October 1994. doi:10.1126/science.7939661. PMID 7939661. Bibcode: 1994Sci...266..271J.
- ↑ Mohammad Afzal (January 1983). "Consanguinity effects on Intelligence Quotient and neonatal behaviors of Ansari muslim children". https://www.researchgate.net/publication/263849940.
- ↑ Sommer, S. (2005). "The importance of immune gene variability (MHC) in evolutionary ecology and conservation". Frontiers in Zoology 2: 16. doi:10.1186/1742-9994-2-16. PMID 16242022.
- ↑ "The genetics of inbreeding depression". Nat. Rev. Genet. 10 (11): 783–96. 2009. doi:10.1038/nrg2664. PMID 19834483.
- ↑ "The molecular basis of the evolution of sex". Molecular Genetics of Development. Advances in Genetics. 24. 1987. 323–70. doi:10.1016/s0065-2660(08)60012-7. ISBN 9780120176243.
- ↑ Michod, R.E. (1994). "Eros and Evolution: A Natural Philosophy of Sex" Addison-Wesley Publishing Company, Reading, Massachusetts. ISBN:978-0201442328
- ↑ "Inbreeding depression in the wild". Heredity 83 (3): 260–270. 1999. doi:10.1038/sj.hdy.6885530. PMID 10504423.
- ↑ "Genetic basis for species vulnerability in the cheetah". Science 227 (4693): 1428–1434. 1985. doi:10.1126/science.2983425. PMID 2983425. Bibcode: 1985Sci...227.1428O.
- ↑ 12.00 12.01 12.02 12.03 12.04 12.05 12.06 12.07 12.08 12.09 12.10 "Inbreeding avoidance in animals". Trends Ecol Evol 11 (5): 201–206. 1996. doi:10.1016/0169-5347(96)10028-8. PMID 21237809.
- ↑ 13.0 13.1 Simmons, L.W. (1989) Kin recognition and its influence on mating preferences of the field cricket, Gryffus bimaculatus (de Geer), Anim. Behav. 38,68-77
- ↑ 14.0 14.1 14.2 Krackow, S. and Matuschak, B. (1991) Mate choice for non-siblings in wild house mice: evidence from a choice test and a reproductive test, Ethology 88,99-108
- ↑ Bollinger, E.K. et al. (1991) Avoidance of inbreeding in the meadow vole (Microtus pennsylvanicus), .I Mammal. 72, 419-421
- ↑ Keane, B. (1990) The effect of relatedness on reproductive success and mate choice in the white-footed mouse, Peromyscus leucopus, Anim. Behav. 39,264-273
- ↑ Wolf, A.P. Westermarck Redivivus. Annual Review of Anthropology 22: 157-175, 1993
- ↑ Holmes WG, Sherman PW (1982) The ontogeny of kin recognition in two species of ground squirrels. American Zoologist, 22,491?517.
- ↑ Pusey, A.E. (1990) Mechanisms of inbreeding avoidance in nonhuman primates, in Pedophilia: Biosocial Dimensions (Feirman, J.R., ed.), pp. 201-220, Springer-Verlag
- ↑ Mateo JM, Johnston RE (2000) Kin recognition and the ‘armpit effect’: evidence of self-referent phenotype matching. Proceedings of the Royal Society of London. Series B, Biological Sciences, 267, 695?700.
- ↑ Jerram L. Brown and Amy Eklund The American Naturalist Vol. 143, No. 3 (Mar., 1994), pp. 435-461 Published by: The University of Chicago Press
- ↑ "Communal nesting patterns in mice implicate MHC genes in kin recognition". Nature 360 (6404): 581–583. 1992. doi:10.1038/360581a0. PMID 1461279. Bibcode: 1992Natur.360..581M.
- ↑ "Possible olfaction-based mechanisms in human kin recognition and inbreeding avoidance". J Exp Child Psychol 85 (3): 279–95. 2003. doi:10.1016/s0022-0965(03)00061-4. PMID 12810039.
- ↑ "Gametic interactions promote inbreeding avoidance in house mice". Ecol. Lett. 18 (9): 937–43. 2015. doi:10.1111/ele.12471. PMID 26154782.
- ↑ "Evidence for inbreeding depression and post-pollination selection against inbreeding in the dioecious plant Silene latifolia". Heredity (Edinb) 102 (2): 101–12. 2009. doi:10.1038/hdy.2008.86. PMID 18698334.
- ↑ "Inbreeding avoidance mechanisms: dispersal dynamics in cooperatively breeding southern pied babblers". Journal of Animal Ecology 81 (4): 876–883. 2012. doi:10.1111/j.1365-2656.2012.01983.x. PMID 22471769.
- ↑ 27.0 27.1 27.2 "Dispersal and inbreeding avoidance". The American Naturalist 154 (3): 282–292. 1999. doi:10.1086/303236. PMID 10506544.
- ↑ 28.0 28.1 28.2 28.3 "Sex-biased dispersal and inbreeding avoidance in birds and mammals". Trends in Ecology & Evolution 2 (10): 295–299. 1987. doi:10.1016/0169-5347(87)90081-4. PMID 21227869.
- ↑ "Inbreeding avoidance through kin recognition: choosy females boost male dispersal". The American Naturalist 162 (5): 638–652. 2003. doi:10.1086/378823. PMID 14618541. https://serval.unil.ch/notice/serval:BIB_00B8BDB73912.
- ↑ 30.0 30.1 "Female transfer and inbreeding avoidance in social mammals". Nature 337 (6202): 70–72. 1989. doi:10.1038/337070a0. PMID 2909891. Bibcode: 1989Natur.337...70C.
- ↑ "Inbreeding avoidance in chimpanzees". Animal Behaviour 28 (2): 543–552. 1980. doi:10.1016/s0003-3472(80)80063-7.
- ↑ "Inbreeding avoidance and male-biased natal dispersal in Antechinus spp. (Marsupialia: Dasyuridae)". Animal Behaviour 33 (3): 908–915. 1985. doi:10.1016/s0003-3472(85)80025-7.
- ↑ 33.0 33.1 "Reproductive suppression and inbreeding avoidance in wild populations of co-operatively breeding meerkats Suricata suricatta". Behav. Ecol. Sociobiol. 48 (6): 471–477. 2000b. doi:10.1007/s002650000249.
- ↑ 34.0 34.1 "Extra-pair paternity in birds: Explaining variation between species and populations". Trends in Ecology and Evolution 13 (2): 52–57. 1998. doi:10.1016/s0169-5347(97)01232-9. PMID 21238200.
- ↑ "Females increase offspring heterozygosity and fitness through extra-pair matings". Nature 425 (6959): 714–7. Oct 2003. doi:10.1038/nature01969. PMID 14562103. Bibcode: 2003Natur.425..714F.
- ↑ Alcock, John. 1998. Animal Behavior. Sixth Edition. 429-519.
- ↑ "A new look at monogamy". Science 281 (5385): 1982–1983. 1998. doi:10.1126/science.281.5385.1982. PMID 9767050.
- ↑ 38.0 38.1 Part, T. (1996). Problems with testing inbreeding avoidance: the case of the collared flycatcher. Evolution, 1625-1630.
- ↑ "Are dispersal and inbreeding avoidance related?". Animal Behaviour 32 (1): 94–112. 1984. doi:10.1016/s0003-3472(84)80328-0.
- ↑ "No evidence of inbreeding avoidance or inbreeding depression in a social carnivore". Behavioral Ecology 7 (4): 480–489. 1996. doi:10.1093/beheco/7.4.480.
 |



