Biology:Eps-Associated RNA element
| eps-associated RNA element | |
|---|---|
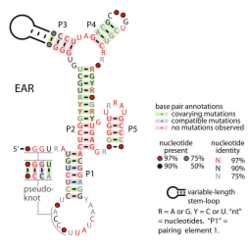 Consensus secondary structure of EAR element RNAs | |
| Identifiers | |
| Symbol | epsC motif |
| Alt. Symbols | EAR |
| Rfam | RF01735 |
| Other data | |
| RNA type | Cis-regulatory element |
| Domain(s) | Bacillales |
| PDB structures | PDBe |
The eps-Associated RNA element (EAR; also called epsC motif) is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales.[1][2] It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon (i.e., between epsB and epsC). Deletion of the EAR element impairs biofilm formation.[1]
The combination of comparative sequence alignments and structural probing data revealed that the secondary structure of the EAR RNA consists of five helical segments (P1-P5) and a pseudoknot[1][2] (see diagram). In general, mutations that disrupt the conserved helical regions eliminate biofilm formation by B. subtilis, whereas mutations in nonconserved, variable regions exhibit no such effect.[1] Thus, the secondary structure of EAR plays an important role for its function in vivo.
At the transcript level, disruption of the EAR motif leads to a significant decrease in the expression of distally located eps genes (epsF-O) without affecting proximal genes (epsC-E).[1] Consistent with this observation, several potential intrinsic terminator hairpins were found inside the epsF coding region.[1] A wild-type EAR element is capable of promoting transcriptional readthrough of these epsF terminator sites. In contrast, mutational disruption of EAR instead permits termination at the epsF terminators.[1] The EAR element also promotes readthrough of heterologous termination sites. From these data it was hypothesized that the EAR element controls eps expression through a processive antitermination mechanism to ensure the complete synthesis of the 16 kilobase operon.[1] Processive antitermination, as opposed to the transcription attenuation mechanism typically used by other cis-acting regulatory RNAs, is a process wherein the transcription elongation complex is altered by accessory factors to become resistant to pausing and termination signals. So far, EAR is the only processive antitermination system specific for Gram-positive bacteria. Further experiments failed to reconstitute EAR-mediated processive antitermination mechanism in vitro or in a Gram-negative heterologous host (E. coli), suggesting that additional B. subtilis-specific factor(s) may also be required for EAR antitermination.[1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 "A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales". Mol Microbiol 76 (3): 559–575. March 2010. doi:10.1111/j.1365-2958.2010.07131.x. PMID 20374491.
- ↑ 2.0 2.1 "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol 11 (3): R31. March 2010. doi:10.1186/gb-2010-11-3-r31. PMID 20230605.
External links
 |

