Earth:Biofilm

- A biofilm is a system that can be adapted internally to environmental conditions by its inhabitants.
- The self-produced matrix of extracellular polymeric substances, which is also referred to as slime, is a polymeric conglomeration generally composed of extracellular biopolymers in various structural forms.[1]
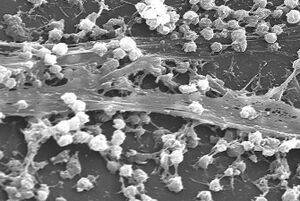
A biofilm is an syntrophic community of microorganisms in which cells stick to each other and often also to a surface.[2][3][4] These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs).[3][4] The cells within the biofilm produce the EPS components, which are typically a polymeric combination of extracellular polysaccharides, proteins, lipids and DNA.[3][4][5] Because they have three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes".[6][7]
Biofilms may form on living (biotic) or non-living (abiotic) surfaces and can be common in natural, industrial, and hospital settings.[4][8] They may constitute a microbiome or be a portion of it. The microbial cells growing in a biofilm are physiologically distinct from planktonic cells of the same organism, which, by contrast, are single cells that may float or swim in a liquid medium.[9] Biofilms can form on the teeth of most animals as dental plaque, where they may cause tooth decay and gum disease.
Microbes form a biofilm in response to a number of different factors,[10] which may include cellular recognition of specific or non-specific attachment sites on a surface, nutritional cues, or in some cases, by exposure of planktonic cells to sub-inhibitory concentrations of antibiotics.[11][12] A cell that switches to the biofilm mode of growth undergoes a phenotypic shift in behavior in which large suites of genes are differentially regulated.[13]
A biofilm may also be considered a hydrogel, which is a complex polymer that contains many times its dry weight in water. Biofilms are not just bacterial slime layers but biological systems; the bacteria organize themselves into a coordinated functional community. Biofilms can attach to a surface such as a tooth or rock, and may include a single species or a diverse group of microorganisms. Subpopulations of cells within the biofilm differentiate to perform various activities for motility, matrix production, and sporulation, supporting the overall success of the biofilm.[14] The biofilm bacteria can share nutrients and are sheltered from harmful factors in the environment, such as desiccation, antibiotics, and a host body's immune system. A biofilm usually begins to form when a free-swimming, planktonic bacterium attaches to a surface.[15][page needed]
Origin and formation
Origin of biofilms
Biofilms are thought to have arisen during primitive Earth as a defense mechanism for prokaryotes, as the conditions at that time were too harsh for their survival. They can be found very early in Earth's fossil records (about 3.25 billion years ago) as both Archaea and Bacteria, and commonly protect prokaryotic cells by providing them with homeostasis, encouraging the development of complex interactions between the cells in the biofilm.[4]
Formation of biofilms
The formation of a biofilm begins with the attachment of free-floating microorganisms to a surface.[9][6] The first colonist bacteria of a biofilm may adhere to the surface initially by the weak van der Waals forces and hydrophobic effects.[16][17] If the colonists are not immediately separated from the surface, they can anchor themselves more permanently using cell adhesion structures such as pili. A unique group of Archaea that inhabit anoxic groundwater have similar structures called hami. Each hamus is a long tube with three hook attachments that are used to attach to each other or to a surface, enabling a community to develop.[18][19] Hyperthermophilic archaeon Pyrobaculum calidifontis produce bundling pili which are homologous to the bacterial TasA filaments, a major component of the extracellular matrix in bacterial biofilms, which contribute to biofilm stability.[20] TasA homologs are encoded by many other archaea, suggesting mechanistic similarities and evolutionary connection between bacterial and archaeal biofilms.[20]
Hydrophobicity can also affect the ability of bacteria to form biofilms. Bacteria with increased hydrophobicity have reduced repulsion between the substratum and the bacterium.[21] Some bacteria species are not able to attach to a surface on their own successfully due to their limited motility but are instead able to anchor themselves to the matrix or directly to other, earlier bacteria colonists. Non-motile bacteria cannot recognize surfaces or aggregate together as easily as motile bacteria.[21]
During surface colonization bacteria cells are able to communicate using quorum sensing (QS) products such as N-acyl homoserine lactone (AHL). Once colonization has begun, the biofilm grows by a combination of cell division and recruitment. Polysaccharide matrices typically enclose bacterial biofilms. The matrix exopolysaccharides can trap QS autoinducers within the biofilm to prevent predator detection and ensure bacterial survival.[22] In addition to the polysaccharides, these matrices may also contain material from the surrounding environment, including but not limited to minerals, soil particles, and blood components, such as erythrocytes and fibrin.[21] The final stage of biofilm formation is known as development, and is the stage in which the biofilm is established and may only change in shape and size.
The development of a biofilm may allow for an aggregate cell colony (or colonies) to be increasingly tolerant[23] or resistant to antibiotics. Cell-cell communication or quorum sensing has been shown to be involved in the formation of biofilm in several bacterial species.[24]
Development

Biofilms are the product of a microbial developmental process.[27] The process is summarized by five major stages of biofilm development, as shown in the diagram below:[28]

Dispersal
Dispersal of cells from the biofilm colony is an essential stage of the biofilm life cycle. Dispersal enables biofilms to spread and colonize new surfaces. Enzymes that degrade the biofilm extracellular matrix, such as dispersin B and deoxyribonuclease, may contribute to biofilm dispersal.[29][30] Enzymes that degrade the biofilm matrix may be useful as anti-biofilm agents.[31][32] Evidence has shown that a fatty acid messenger, cis-2-decenoic acid, is capable of inducing dispersion and inhibiting growth of biofilm colonies. Secreted by Pseudomonas aeruginosa, this compound induces cyclo heteromorphic cells in several species of bacteria and the yeast Candida albicans.[33] Nitric oxide has also been shown to trigger the dispersal of biofilms of several bacteria species[34][35] at sub-toxic concentrations. Nitric oxide has potential as a treatment for patients that have chronic infections caused by biofilms.[36]
It was generally assumed that cells dispersed from biofilms immediately go into the planktonic growth phase. However, studies have shown that the physiology of dispersed cells from Pseudomonas aeruginosa biofilms is highly different from that of planktonic and biofilm cells.[37][38] Hence, the dispersal process is a unique stage during the transition from biofilm to planktonic lifestyle in bacteria. Dispersed cells are found to be highly virulent against macrophages and Caenorhabditis elegans, but highly sensitive towards iron stress, as compared with planktonic cells.[37]
Furthermore, Pseudomonas aeruginosa biofilms undergo distinct spatiotemporal dynamics during biofilm dispersal or disassembly, with contrasting consequences in recolonization and disease dissemination.[39] Biofilm dispersal induced bacteria to activate dispersal genes to actively depart from biofilms as single cells at consistent velocities but could not recolonize fresh surfaces. In contrast, biofilm disassembly by degradation of a biofilm exopolysaccharide released immotile aggregates at high initial velocities, enabling the bacteria to recolonize fresh surfaces and cause infections in the hosts efficiently. Hence, biofilm dispersal is more complex than previously thought, where bacterial populations adopting distinct behavior after biofilm departure may be the key to survival of bacterial species and dissemination of diseases.

Properties
Biofilms are usually found on solid substrates submerged in or exposed to an aqueous solution, although they can form as floating mats on liquid surfaces and also on the surface of leaves, particularly in high humidity climates. Given sufficient resources for growth, a biofilm will quickly grow to be macroscopic (visible to the naked eye). Biofilms can contain many different types of microorganism, e.g. bacteria, archaea, protozoa, fungi and algae; each group performs specialized metabolic functions. However, some organisms will form single-species films under certain conditions. The social structure (cooperation/competition) within a biofilm depends highly on the different species present.[40]
Extracellular matrix

The EPS matrix consists of exopolysaccharides, proteins and nucleic acids.[41][42][43] A large proportion of the EPS is more or less strongly hydrated, however, hydrophobic EPS also occur; one example is cellulose[44] which is produced by a range of microorganisms. This matrix encases the cells within it and facilitates communication among them through biochemical signals as well as gene exchange. The EPS matrix also traps extracellular enzymes and keeps them in close proximity to the cells. Thus, the matrix represents an external digestion system and allows for stable synergistic microconsortia of different species.[45] Some biofilms have been found to contain water channels that help distribute nutrients and signalling molecules.[46] This matrix is strong enough that under certain conditions, biofilms can become fossilized (stromatolites).
Bacteria living in a biofilm usually have significantly different properties from free-floating bacteria of the same species, as the dense and protected environment of the film allows them to cooperate and interact in various ways.[47] One benefit of this environment is increased resistance to detergents and antibiotics, as the dense extracellular matrix and the outer layer of cells protect the interior of the community.[48][49] In some cases antibiotic resistance can be increased up to 5,000 times.[50] Lateral gene transfer is often facilitated within bacterial and archaeal biofilms[51] and can leads to a more stable biofilm structure.[52] Extracellular DNA is a major structural component of many different microbial biofilms.[53] Enzymatic degradation of extracellular DNA can weaken the biofilm structure and release microbial cells from the surface.
However, biofilms are not always less susceptible to antibiotics. For instance, the biofilm form of Pseudomonas aeruginosa has no greater resistance to antimicrobials than do stationary-phase planktonic cells, although when the biofilm is compared to logarithmic-phase planktonic cells, the biofilm does have greater resistance to antimicrobials. This resistance to antibiotics in both stationary-phase cells and biofilms may be due to the presence of persister cells.[54]
Habitats


Biofilms are ubiquitous in organic life. Nearly every species of microorganism have mechanisms by which they can adhere to surfaces and to each other. Biofilms will form on virtually every non-shedding surface in non-sterile aqueous or humid environments. Biofilms can grow in the most extreme environments: from, for example, the extremely hot, briny waters of hot springs ranging from very acidic to very alkaline, to frozen glaciers.
Biofilms can be found on rocks and pebbles at the bottoms of most streams or rivers and often form on the surfaces of stagnant pools of water. Biofilms are important components of food chains in rivers and streams and are grazed by the aquatic invertebrates upon which many fish feed. Biofilms are found on the surface of and inside plants. They can either contribute to crop disease or, as in the case of nitrogen-fixing rhizobia on root nodules, exist symbiotically with the plant.[55] Examples of crop diseases related to biofilms include citrus canker, Pierce's disease of grapes, and bacterial spot of plants such as peppers and tomatoes.[56]
Percolating filters
Percolating filters in sewage treatment works are highly effective removers of pollutants from settled sewage liquor. They work by trickling the liquid over a bed of hard material which is designed to have a very large surface area. A complex biofilm develops on the surface of the medium which absorbs, adsorbs and metabolises the pollutants. The biofilm grows rapidly and when it becomes too thick to retain its grip on the media it washes off and is replaced by newly grown film. The washed off ("sloughed" off) film is settled out of the liquid stream to leave a highly purified effluent.[57]
Slow sand filter
Slow sand filters are used in water purification for treating raw water to produce a potable product. They work through the formation of a biofilm called the hypogeal layer or Schmutzdecke in the top few millimetres of the fine sand layer. The Schmutzdecke is formed in the first 10–20 days of operation[58] and consists of bacteria, fungi, protozoa, rotifera and a range of aquatic insect larvae. As an epigeal biofilm ages, more algae tend to develop and larger aquatic organisms may be present including some bryozoa, snails and annelid worms. The surface biofilm is the layer that provides the effective purification in potable water treatment, the underlying sand providing the support medium for this biological treatment layer. As water passes through the hypogeal layer, particles of foreign matter are trapped in the mucilaginous matrix and soluble organic material is adsorbed. The contaminants are metabolised by the bacteria, fungi and protozoa. The water produced from an exemplary slow sand filter is of excellent quality with 90–99% bacterial cell count reduction.[59]
Rhizosphere
Plant-beneficial microbes can be categorized as plant growth-promoting rhizobacteria.[60] These plant growth-promoters colonize the roots of plants, and provide a wide range of beneficial functions for their host including nitrogen fixation, pathogen suppression, anti-fungal properties, and the breakdown of organic materials.[61] One of these functions is the defense against pathogenic, soil-borne bacteria and fungi by way of induced systemic resistance (ISR)[62] or induced systemic responses triggered by pathogenic microbes (pathogen-induced systemic acquired resistance).[63] Plant exudates act as chemical signals for host specific bacteria to colonize.[64] Rhizobacteria colonization steps include attractions, recognition, adherence, colonization, and growth.[61] Bacteria that have been shown to be beneficial and form biofilms include Bacillus, Pseudomonas, and Azospirillum.[65][66] Biofilms in the rhizosphere often result in pathogen or plant induced systemic resistances. Molecular properties on the surface of the bacterium cause an immune response in the plant host.[64] These microbe associated molecules interact with receptors on the surface of plant cells, and activate a biochemical response that is thought to include several different genes at a number of loci.[64] Several other signaling molecules have been linked to both induced systemic responses and pathogen-induced systemic responses, such as jasmonic acid and ethylene.[61] Cell envelope components such as bacterial flagella and lipopolysaccharides, which are recognized by plant cells as components of pathogens.[67] Certain iron metabolites produced by Pseudomonas have also been shown to create an induced systemic response.[64] This function of the biofilm helps plants build stronger resistance to pathogens.
Plants that have been colonized by PGPR forming a biofilm have gained systemic resistances and are primed for defense against pathogens. This means that the genes necessary for the production of proteins that work towards defending the plant against pathogens have been expressed, and the plant has a "stockpile" of compounds to release to fight off pathogens.[64] A primed defense system is much faster in responding to pathogen induced infection, and may be able to deflect pathogens before they are able to establish themselves.[68] Plants increase the production of lignin, reinforcing cell walls and making it difficult for pathogens to penetrate into the cell, while also cutting off nutrients to already infected cells, effectively halting the invasion.[61] They produce antimicrobial compounds such as phytoalexins, chitinases, and proteinase inhibitors, which prevent the growth of pathogens.[63] These functions of disease suppression and pathogen resistance ultimately lead to an increase in agricultural production and a decrease in the use of chemical pesticides, herbicides, and fungicides because there is a reduced amount of crop loss due to disease.[69] Induced systemic resistance and pathogen-induced systemic acquired resistance are both potential functions of biofilms in the rhizosphere, and should be taken into consideration when applied to new age agricultural practices because of their effect on disease suppression without the use of dangerous chemicals.
Mammalian gut
Studies in 2003 discovered that the immune system supports biofilm development in the large intestine. This was supported mainly with the fact that the two most abundantly produced molecules by the immune system also support biofilm production and are associated with the biofilms developed in the gut. This is especially important because the appendix holds a mass amount of these bacterial biofilms.[70] This discovery helps to distinguish the possible function of the appendix and the idea that the appendix can help reinoculate the gut with good gut flora. However, modified or disrupted states of biofilms in the gut have been connected to diseases such as inflammatory bowel disease and colorectal cancer.[71]
Human environment
In the human environment, biofilms can grow in showers very easily since they provide a moist and warm environment for them to thrive. They can form inside water and sewage pipes and cause clogging and corrosion. On floors and counters, they can make sanitation difficult in food preparation areas. In soil, they can cause bioclogging. In cooling- or heating-water systems, they are known to reduce heat transfer.[72] Biofilms in marine engineering systems, such as pipelines of the offshore oil and gas industry,[73] can lead to substantial corrosion problems. Corrosion is mainly due to abiotic factors; however, at least 20% of corrosion is caused by microorganisms that are attached to the metal subsurface (i.e., microbially influenced corrosion).
Ship fouling
Bacterial adhesion to boat hulls serves as the foundation for biofouling of seagoing vessels. Once a film of bacteria forms, it is easier for other marine organisms such as barnacles to attach. Such fouling can reduce maximum vessel speed by up to 20%, prolonging voyages and consuming fuel. Time in dry dock for refitting and repainting reduces the productivity of shipping assets, and the useful life of ships is also reduced due to corrosion and mechanical removal (scraping) of marine organisms from ships' hulls.
Stromatolites
Stromatolites are layered accretionary structures formed in shallow water by the trapping, binding and cementation of sedimentary grains by microbial biofilms, especially of cyanobacteria. Stromatolites include some of the most ancient records of life on Earth, and are still forming today.
Dental plaque
Within the human body, biofilms are present on the teeth as dental plaque, where they may cause tooth decay and gum disease. These biofilms can either be in an uncalcified state that can be removed by dental instruments, or a calcified state which is more difficult to remove. Removal techniques can also include antimicrobials.[74]
Dental plaque is an oral biofilm that adheres to the teeth and consists of many species of both bacteria and fungi (such as Streptococcus mutans and Candida albicans), embedded in salivary polymers and microbial extracellular products. The accumulation of microorganisms subjects the teeth and gingival tissues to high concentrations of bacterial metabolites which results in dental disease.[75] Biofilm on the surface of teeth is frequently subject to oxidative stress[76] and acid stress.[77] Dietary carbohydrates can cause a dramatic decrease in pH in oral biofilms to values of 4 and below (acid stress).[77] A pH of 4 at body temperature of 37 °C causes depurination of DNA, leaving apurinic (AP) sites in DNA,[78] especially loss of guanine.[79]
Dental plaque biofilm can result in dental caries if it is allowed to develop over time. An ecologic shift away from balanced populations within the dental biofilm is driven by certain (cariogenic) microbiological populations beginning to dominate when the environment favors them. The shift to an acidogenic, aciduric, and cariogenic microbiological population develops and is maintained by frequent consumption of fermentable dietary carbohydrate. The resulting activity shift in the biofilm (and resulting acid production within the biofilm, at the tooth surface) is associated with an imbalance of demineralization over remineralization, leading to net mineral loss within dental hard tissues (enamel and then dentin), the symptom being a carious lesion, or cavity. By preventing the dental plaque biofilm from maturing or by returning it back to a non-cariogenic state, dental caries can be prevented and arrested.[80][81] This can be achieved through the behavioral step of reducing the supply of fermentable carbohydrates (i.e. sugar intake) and frequent removal of the biofilm (i.e., toothbrushing).[80]
Intercellular communication
A peptide pheromone quorum sensing signaling system in S. mutans includes the competence stimulating peptide (CSP) that controls genetic competence.[82][83] Genetic competence is the ability of a cell to take up DNA released by another cell. Competence can lead to genetic transformation, a form of sexual interaction, favored under conditions of high cell density and/or stress where there is maximal opportunity for interaction between the competent cell and the DNA released from nearby donor cells. This system is optimally expressed when S. mutans cells reside in an actively growing biofilm. Biofilm grown S. mutans cells are genetically transformed at a rate 10- to 600-fold higher than S. mutans growing as free-floating planktonic cells suspended in liquid.[82]
When the biofilm, containing S. mutans and related oral streptococci, is subjected to acid stress, the competence regulon is induced, leading to resistance to being killed by acid.[77] As pointed out by Michod et al., transformation in bacterial pathogens likely provides for effective and efficient recombinational repair of DNA damages.[84] It appears that S. mutans can survive the frequent acid stress in oral biofilms, in part, through the recombinational repair provided by competence and transformation.
Predator-prey interactions
Predator-prey interactions between biofilms and bacterivores, such as the soil-dwelling nematode Caenorhabditis elegans, had been extensively studied. Via the production of sticky matrix and formation of aggregates, Yersinia pestis biofilms can prevent feeding by obstructing the mouth of C. elegans.[85] Moreover, Pseudomonas aeruginosa biofilms can impede the slithering motility of C. elegans, termed as 'quagmire phenotype', resulting in trapping of C. elegans within the biofilms and preventing the exploration of nematodes to feed on susceptible biofilms.[86] This significantly reduced the ability of predator to feed and reproduce, thereby promoting the survival of biofilms. Pseudomonas aeruginosa biofilms can also mask their chemical signatures, where they reduced the diffusion of quorum sensing molecules into the environment and prevented the detection of C. elegans.[87]
Taxonomic diversity
Many different bacteria form biofilms, including gram-positive (e.g. Bacillus spp, Listeria monocytogenes, Staphylococcus spp, and lactic acid bacteria, including Lactobacillus plantarum and Lactococcus lactis) and gram-negative species (e.g. Escherichia coli, or Pseudomonas aeruginosa).[88] Cyanobacteria also form biofilms in aquatic environments.[89]
Biofilms are formed by bacteria that colonize plants, e.g. Pseudomonas putida, Pseudomonas fluorescens, and related pseudomonads which are common plant-associated bacteria found on leaves, roots, and in the soil, and the majority of their natural isolates form biofilms.[90] Several nitrogen-fixing symbionts of legumes such as Rhizobium leguminosarum and Sinorhizobium meliloti form biofilms on legume roots and other inert surfaces.[90]
Along with bacteria, biofilms are also generated by archaea[51] and by a range of eukaryotic organisms, including fungi e.g. Cryptococcus laurentii[91] and microalgae. Among microalgae, one of the main progenitors of biofilms are diatoms, which colonise both fresh and marine environments worldwide.[92][93]
For other species in disease-associated biofilms and biofilms arising from eukaryotes, see below.
Infectious diseases
Biofilms have been found to be involved in a wide variety of microbial infections in the body, by one estimate 80% of all infections.[94] Infectious processes in which biofilms have been implicated include common problems such as bacterial vaginosis, urinary tract infections, catheter infections, middle-ear infections, formation of dental plaque,[95] gingivitis, coating contact lenses,[96] and less common but more lethal processes such as endocarditis, infections in cystic fibrosis, and infections of permanent indwelling devices such as joint prostheses, heart valves, and intervertebral disc.[97][98][99] The first visual evidence of a biofilm was recorded after spine surgery.[100] It was found that in the absence of clinical presentation of infection, impregnated bacteria could form a biofilm around an implant, and this biofilm can remain undetected via contemporary diagnostic methods, including swabbing. Implant biofilm is frequently present in "aseptic" pseudarthrosis cases.[100][101][102] Furthermore, it has been noted that bacterial biofilms may impair cutaneous wound healing and reduce topical antibacterial efficiency in healing or treating infected skin wounds.[103] The diversity of P. aeruginosa cells within a biofilm is thought to make it harder to treat the infected lungs of people with cystic fibrosis.[14] Early detection of biofilms in wounds is crucial to successful chronic wound management. Although many techniques have developed to identify planktonic bacteria in viable wounds, few have been able to quickly and accurately identify bacterial biofilms. Future studies are needed to find means of identifying and monitoring biofilm colonization at the bedside to permit timely initiation of treatment.[104]
It has been shown that biofilms are present on the removed tissue of 80% of patients undergoing surgery for chronic sinusitis. The patients with biofilms were shown to have been denuded of cilia and goblet cells, unlike the controls without biofilms who had normal cilia and goblet cell morphology.[105] Biofilms were also found on samples from two of 10 healthy controls mentioned. The species of bacteria from intraoperative cultures did not correspond to the bacteria species in the biofilm on the respective patient's tissue. In other words, the cultures were negative though the bacteria were present.[106] New staining techniques are being developed to differentiate bacterial cells growing in living animals, e.g. from tissues with allergy-inflammations.[107]
Research has shown that sub-therapeutic levels of β-lactam antibiotics induce biofilm formation in Staphylococcus aureus. This sub-therapeutic level of antibiotic may result from the use of antibiotics as growth promoters in agriculture, or during the normal course of antibiotic therapy. The biofilm formation induced by low-level methicillin was inhibited by DNase, suggesting that the sub-therapeutic levels of antibiotic also induce extracellular DNA release.[108] Moreover, from an evolutionary point of view, the creation of the tragedy of the commons in pathogenic microbes may provide advanced therapeutic ways for chronic infections caused by biofilms via genetically engineered invasive cheaters who can invade wild-types 'cooperators' of pathogenic bacteria until cooperator populations go to extinction or overall population 'cooperators and cheaters ' go to extinction.[109]
Pseudomonas aeruginosa
P. aeruginosa represents a commonly used biofilm model organism since it is involved in different types of biofilm-associated chronic infections.[41] Examples of such infections include chronic wounds, chronic otitis media, chronic prostatitis and chronic lung infections in cystic fibrosis (CF) patients. About 80% of CF patients have chronic lung infection, caused mainly by P. aeruginosa growing in a non-surface attached biofilms surround by PMN.[110] The infection remains present despite aggressive antibiotic therapy and is a common cause of death in CF patients due to constant inflammatory damage to the lungs.[41] In patients with CF, one therapy for treating early biofilm development is to employ DNase to structurally weaken the biofilm.[5][111]
Biofilm formation of P. aeruginosa, along with other bacteria, is found in 90% of chronic wound infections, which leads to poor healing and high cost of treatment estimated at more than US$25 billion every year in the United States .[112] In order to minimize the P. aeruginosa infection, host epithelial cells secrete antimicrobial peptides, such as lactoferrin, to prevent the formation of the biofilms.[113]
Streptococcus pneumoniae
Streptococcus pneumoniae is the main cause of community-acquired pneumonia and meningitis in children and the elderly, and of sepsis in HIV-infected persons. When S. pneumoniae grows in biofilms, genes are specifically expressed that respond to oxidative stress and induce competence.[114] Formation of a biofilm depends on competence stimulating peptide (CSP). CSP also functions as a quorum-sensing peptide. It not only induces biofilm formation, but also increases virulence in pneumonia and meningitis.
It has been proposed that competence development and biofilm formation is an adaptation of S. pneumoniae to survive the defenses of the host.[84] In particular, the host's polymorphonuclear leukocytes produce an oxidative burst to defend against the invading bacteria, and this response can kill bacteria by damaging their DNA. Competent S. pneumoniae in a biofilm have the survival advantage that they can more easily take up transforming DNA from nearby cells in the biofilm to use for recombinational repair of oxidative damages in their DNA. Competent S. pneumoniae can also secrete an enzyme (murein hydrolase) that destroys non-competent cells (fratricide) causing DNA to be released into the surrounding medium for potential use by the competent cells.[115]
The insect antimicrobial peptide cecropin A can destroy planktonic and sessile biofilm-forming uropathogenic E. coli cells, either alone or when combined with the antibiotic nalidixic acid, synergistically clearing infection in vivo (in the insect host Galleria mellonella) without off-target cytotoxicity. The multi-target mechanism of action involves outer membrane permeabilization followed by biofilm disruption triggered by the inhibition of efflux pump activity and interactions with extracellular and intracellular nucleic acids.[116]
Escherichia coli
Escherichia coli biofilms are responsible for many intestinal infectious diseases.[117] The Extraintestinal group of E. coli (ExPEC) is the dominant bacterial group that attacks the urinary system, which leads to urinary tract infections.[118] The biofilm formation of these pathogenic E. coli is hard to eradicate due to the complexity of its aggregation structure, and it has a significant contribution to developing aggressive medical complications, increase in hospitalization rate, and cost of treatment.[119][120] The development of E. coli biofilm is a common leading cause of urinary tract infections (UTI) in hospitals through its contribution to developing medical device-associated infections. Catheter-associated urinary tract infections (CAUTI) represent the most common hospital-acquired infection due to the formation of the pathogenic E. coli biofilm inside the catheters.[121]
Staphylococcus aureus
Staphylococcus aureus pathogen can attack skin and lungs, leading to skin infection and pneumonia.[122][123] Moreover, the biofilm infections network of S. aureus plays a critical role in preventing immune cells, such as macrophages from eliminating and destroying bacterial cells.[124] Furthermore, biofilm formation by bacteria, such as S. aureus, not only develops resistance against antibiotic medication but also develop internal resistance toward antimicrobial peptides (AMPs), leading to preventing the inhibition of the pathogen and maintaining its survival.[125]
Uses and impact
In medicine
It is suggested that around two-thirds of bacterial infections in humans involve biofilms.[50][126] Infections associated with the biofilm growth usually are challenging to eradicate.[127] This is mostly due to the fact that mature biofilms display antimicrobial tolerance, and immune response evasions.[128][41] Biofilms often form on the inert surfaces of implanted devices such as catheters, prosthetic cardiac valves and intrauterine devices.[129] Some of the most difficult infections to treat are those associated with the use of medical devices.[50][101]
The rapidly expanding worldwide industry for biomedical devices and tissue engineering related products is already at $180 billion per year, yet this industry continues to suffer from microbial colonization. No matter the sophistication, microbial infections can develop on all medical devices and tissue engineering constructs.[128] 60-70% of hospital-acquired infections are associated with the implantation of a biomedical device.[128] This leads to 2 million cases annually in the U.S., costing the healthcare system over $5 billion in additional healthcare expenses.[128]
The level of antibiotic resistance in a biofilm is much greater than that of non-biofilm bacteria, and can be as much as 5,000 times greater.[50] The extracellular matrix of biofilm is considered one of the leading factors that can reduce the penetration of antibiotics into a biofilm structure and contributes to antibiotic resistance.[130] Further, it has been demonstrated that the evolution of resistance to antibiotics may be affected by the biofilm lifestyle.[131] Bacteriophage therapy can disperse the biofilm generated by antibiotic-resistant bacteria.[132]
It has been shown that the introduction of a small current of electricity to the liquid surrounding a biofilm, together with small amounts of antibiotic can reduce the level of antibiotic resistance to levels of non-biofilm bacteria. This is termed the bioelectric effect.[50][133] The application of a small DC current on its own can cause a biofilm to detach from its surface.[50] A study showed that the type of current used made no difference to the bioelectric effect.[133]
In industry
Biofilms can also be harnessed for constructive purposes. For example, many sewage treatment plants include a secondary treatment stage in which waste water passes over biofilms grown on filters, which extract and digest organic compounds. In such biofilms, bacteria are mainly responsible for removal of organic matter (BOD), while protozoa and rotifers are mainly responsible for removal of suspended solids (SS), including pathogens and other microorganisms. Slow sand filters rely on biofilm development in the same way to filter surface water from lake, spring or river sources for drinking purposes. What is regarded as clean water is effectively a waste material to these microcellular organisms. Biofilms can help eliminate petroleum oil from contaminated oceans or marine systems. The oil is eliminated by the hydrocarbon-degrading activities of communities of hydrocarbonoclastic bacteria (HCB).[134] Biofilms are used in microbial fuel cells (MFCs) to generate electricity from a variety of starting materials, including complex organic waste and renewable biomass.[8][135][136] Biofilms are also relevant for the improvement of metal dissolution in bioleaching industry,[137] and aggregation of microplastics pollutants for convenient removal from the environment.[138][139]
Food industry
Biofilms have become problematic in several food industries due to the ability to form on plants and during industrial processes.[140] Bacteria can survive long periods of time in water, animal manure, and soil, causing biofilm formation on plants or in the processing equipment.[141] The buildup of biofilms can affect the heat flow across a surface and increase surface corrosion and frictional resistance of fluids.[142] These can lead to a loss of energy in a system and overall loss of products.[142] Along with economic problems, biofilm formation on food poses a health risk to consumers due to the ability to make the food more resistant to disinfectants[140] As a result, from 1996 to 2010 the Centers for Disease Control and Prevention estimated 48 million foodborne illnesses per year.[140] Biofilms have been connected to about 80% of bacterial infections in the United States.[140]
In produce, microorganisms attach to the surfaces and biofilms develop internally.[140] During the washing process, biofilms resist sanitization and allow bacteria to spread across the produce,[140] especially via kitchen utensils.[143] This problem is also found in ready-to-eat foods, because the foods go through limited cleaning procedures before consumption[140] Due to the perishability of dairy products and limitations in cleaning procedures, resulting in the buildup of bacteria, dairy is susceptible to biofilm formation and contamination.[140][142] The bacteria can spoil the products more readily and contaminated products pose a health risk to consumers. One species of bacteria that can be found in various industries and is a major cause of foodborne disease is Salmonella.[144] Large amounts of Salmonella contamination can be found in the poultry processing industry as about 50% of Salmonella strains can produce biofilms on poultry farms.[140] Salmonella increases the risk of foodborne illnesses when the poultry products are not cleaned and cooked correctly. Salmonella is also found in the seafood industry where biofilms form from seafood borne pathogens on the seafood itself as well as in water.[144] Shrimp products are commonly affected by Salmonella because of unhygienic processing and handling techniques[144] The preparation practices of shrimp and other seafood products can allow for bacteria buildup on the products.[144]
New forms of cleaning procedures are being tested to reduce biofilm formation in these processes which will lead to safer and more productive food processing industries. These new forms of cleaning procedures also have a profound effect on the environment, often releasing toxic gases into the groundwater reservoirs.[142] As a response to the aggressive methods employed in controlling biofilm formation, there are a number of novel technologies and chemicals under investigation that can prevent either the proliferation or adhesion of biofilm-secreting microbes. Latest proposed biomolecules presenting marked anti-biofilm activity include a range of metabolites such as bacterial rhamnolipids[145] and even plant-[146] and animal-derived alkaloids.[147]
In aquaculture

In shellfish and algal aquaculture, biofouling microbial species tend to block nets and cages and ultimately outcompete the farmed species for space and food.[148] Bacterial biofilms start the colonization process by creating microenvironments that are more favorable for biofouling species. In the marine environment, biofilms could reduce the hydrodynamic efficiency of ships and propellers, lead to pipeline blockage and sensor malfunction, and increase the weight of appliances deployed in seawater.[149] Numerous studies have shown that biofilm can be a reservoir for potentially pathogenic bacteria in freshwater aquaculture.[150][151][152][153] Moreover, biofilms are important in establishing infections on the fish.[154] As mentioned previously, biofilms can be difficult to eliminate even when antibiotics or chemicals are used in high doses.[155][156] The role that biofilm plays as reservoirs of bacterial fish pathogens has not been explored in detail but it certainly deserves to be studied.
Eukaryotic
Along with bacteria, biofilms are often initiated and produced by eukaryotic microbes. The biofilms produced by eukaryotes is usually occupied by bacteria and other eukaryotes alike, however the surface is cultivated and EPS is secreted initially by the eukaryote.[91][92][157] Both fungi and microalgae are known to form biofilms in such a way. Biofilms of fungal origin are important aspects of human infection and fungal pathogenicity, as the fungal infection is more resistant to antifungals.[158][159]
In the environment, fungal biofilms are an area of ongoing research. One key area of research is fungal biofilms on plants. For example, in the soil, plant associated fungi including mycorrhiza have been shown to decompose organic matter and protect plants from bacterial pathogens.[160]
Biofilms in aquatic environments are often founded by diatoms. The exact purpose of these biofilms is unknown, however there is evidence that the EPS produced by diatoms facilitates both cold and salinity stress.[93][161] These eukaryotes interact with a diverse range of other organisms within a region known as the phycosphere, but importantly are the bacteria associated with diatoms, as it has been shown that although diatoms excrete EPS, they only do so when interacting with certain bacteria species.[162][163]
Horizontal gene transfer
Horizontal gene transfer is the lateral transfer of genetic material between cellular organisms. It happens frequently in prokaryotes, and less frequently in eukaryotes. In bacteria, horizontal gene transfer can occur through transformation (uptake of free floating DNA in the environment), transduction (virus mediated DNA uptake), or conjugation (transfer of DNA between pili structures of two adjacent bacteria).[164] Recent studies have also uncovered other mechanisms, such as membrane vesicle transmission or gene transfer agents.[165] Biofilms promote horizontal gene transfer in a variety of ways.
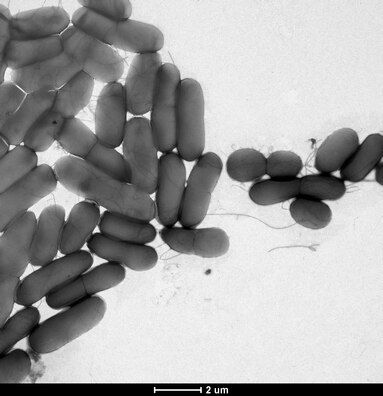
Bacterial conjugation has been shown to accelerate biofilm formation in difficult environment due to the robust connections established by the conjugative pili.[166] These connections can often foster cross-species transfer events due to the diverse heterogeneity of many biofilms. Additionally, biofilms are structurally confined by a polysaccharide matrix, providing the close spatial requirements for conjugation. Transformation is also frequently observed in biofilms. Bacterial autolysis is a key mechanism in biofilm structural regulation, providing an abundant source of competent DNA primed for transformative uptake.[167][165] In some instances, inter-biofilm quorum sensing can enhance the competence of free floating eDNA, further promoting transformation.[165] Stx gene transfer through bacteriophage carriers has been witnessed within biofilms, which suggests that biofilms are also a suitable environment for transduction.[165] Membrane vesicles HGT occurs when released membrane vesicles (containing genetic information) fuse with a recipient bacteria, and release genetic material into the bacteria's cytoplasm.[165] Recent research has revealed that membrane vesicle HGT can promote single-strain biofilm formation, yet the role membrane vesicle HGT plays in the formation of multistrain biofilms is still unknown.[165] GTAs, or gene transfer agents, are phage-like particles produced by the host bacteria and contain random DNA fragments from the host bacteria genome.[165] HGT within biofilms can confer antibiotic resistance or increased pathogenicity across the biofilms' population, promoting biofilm homeostasis.[165]
Examples
Conjugative plasmids may encode biofilm-associated proteins, such as PtgA, PrgB, or PrgC which promote cell adhesion (required for early biofilm formation).[168] Genes encoding type III fimbriae are found in pOLA52 (Klebsiella pneumoniae plasmid) which promote conjugative-pilus-dependent biofilm formation.[168]
Transformation commonly occurs within biofilms. A phenomenon called fratricide can be seen among streptococcal species in which cell-wall degrading enzymes are released, lysing neighboring bacteria and releasing their DNA. This DNA can then be taken up by the surviving bacteria (transformation).[168] Competence stimulating peptides may play an important role in biofilm formation among S. pneumoniae and S. mutans as well.[168] Among V. cholerae, the competence pilus itself promotes cell aggregation through pilus-pilus interactions at the beginning of biofilm formation.[168]
Phage invasion may play a role in biofilm life cycles, lysing bacteria and releasing their eDNA, which strengthens biofilm structures and can be taken up by neighboring bacteria in transformation.[168] Biofilm destruction caused by the E. coli phage Rac and the P. aeruginosa prophage Pf4 causes detachment of cells from the biofilm.[168] Detachment is a biofilm phenomenon which requires more study, but is hypothesized to proliferate the bacterial species that comprise the biofilm.
Membrane vesicle HGT has been witnessed occurring in marine environments, among Neisseria gonorrhoeae, Pseudomonas aeruginosa, Helicobacter pylori, and among many other bacterial species.[168] Even though membrane vesicle HGT has been shown as a contributing factor in biofilm formation, research is still required to prove that membrane vesicle mediated HGT occurs within biofilms.[165][168] Membrane vesicle HGT has also been shown to modulate phage-bacteria interactions in Bacillus subtilis SPP1 phage-resistant cells (lacking the SPP1 receptor protein). Upon exposure to vesicles containing receptors, transduction of pBT163 (a cat-encoding plasmid) occurs, resulting in the expression of the SPP1 receptor protein, opening the receptive bacteria to future phage infection.[168]
Recent research has shown that the archaeal species H. volcanii has some biofilm phenotypes similar to bacterial biofilms such as differentiation and HGT, which required cell-cell contact and involved formation of cytosolic bridges and cellular fusion events.[169]
Cultivation devices
There is a wide variety of biofilm cultivation devices to mimic natural or industrial environments. Although it is important to consider that the particular experimental platform for biofilm research determines what kind of biofilm is cultivated and the data that can be extracted. These devices can be grouped into the following:[170]
- microtiter plate (MTP) systems and MBEC Assay® [formerly the Calgary Biofilm Device (CBD)]
- BioFilm Ring Test (BRT) or clinical Biofilm Ring Test (cBRT)
- Robbins Device or modified Robbins Device (such as the MPMR-10PMMA or the Bio-inLine Biofilm Reactor)
- Drip Flow Biofilm Reactor®
- rotary devices (such as the CDC Biofilm Reactor®, the Rotating Disk Reactor, the Biofilm Annular Reactor, the Industrial Surfaces Biofilm Reactor, or the Constant Depth Film Fermenter)
- flow chambers or flow cells (such as the Coupon Evaluation Flow Cell, Transmission Flow Cell, and Capillary Flow Cell from BioSurface Technologies)
- microfluidic approaches, such as 3D-bacterial “biofilm-dispersal-then-recolonization” (BDR) microfluidic model [39]
See also
- Biology:Amyloid – Insoluble protein aggregate with a fibrillar morphology
- Biology:Bacterial nanowires – Electrically conductive appendages produced by a number of bacteria
- Biology:Biofilm factory – The use of microbial biofilms for chemical production
- Earth:Biomineralization – Process by which living organisms produce minerals
- Organization:Center for Biofilm Engineering
- Philosophy:Microbial intelligence – Adaptive behavior by microscopic organisms including bacteria and protists
- Biology:Microbial mat – Multi-layered sheet of microorganisms
- Medicine:Phage therapy – Therapeutic use of bacteriophages to treat bacterial infections
- Biology:Phototrophic biofilm – Microbial communities including microorganisms which use light as their energy source
- Earth:Stromatolite – Layered sedimentary structure formed by the growth of bacteria or algae
References
- ↑ "Terminology for biorelated polymers and applications (IUPAC Recommendations 2012)". Pure and Applied Chemistry 84 (2): 377–410. 2012. doi:10.1351/PAC-REC-10-12-04.
- ↑ "Biofilms - an overview | ScienceDirect Topics". https://www.sciencedirect.com/topics/materials-science/biofilms.
- ↑ 3.0 3.1 3.2 "Biofilms". Cold Spring Harbor Perspectives in Biology 2 (7): a000398. July 2010. doi:10.1101/cshperspect.a000398. PMID 20519345.
- ↑ 4.0 4.1 4.2 4.3 4.4 "Bacterial biofilms: from the natural environment to infectious diseases". Nature Reviews. Microbiology 2 (2): 95–108. February 2004. doi:10.1038/nrmicro821. PMID 15040259.
- ↑ 5.0 5.1 "Biofilm Cohesive Strength as a Basis for Biofilm Recalcitrance: Are Bacterial Biofilms Overdesigned?". Microbiology Insights 8 (Suppl 2): 29–32. January 2016. doi:10.4137/MBI.S31444. PMID 26819559.
- ↑ 6.0 6.1 "Biofilm, city of microbes". Journal of Bacteriology 182 (10): 2675–9. May 2000. doi:10.1128/jb.182.10.2675-2679.2000. PMID 10781532.
- ↑ "Building Codes for Bacterial Cities | Quanta Magazine". Quanta Magazine. https://www.quantamagazine.org/building-codes-for-bacterial-cities-20170725/.
- ↑ 8.0 8.1 Microbial Biofilms: Current Research and Applications. Caister Academic Press. 2012. ISBN 978-1-904455-96-7.
- ↑ 9.0 9.1 "Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis". Molecular Microbiology 28 (3): 449–61. May 1998. doi:10.1046/j.1365-2958.1998.00797.x. PMID 9632250.
- ↑ "Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development". Molecular Microbiology 30 (2): 295–304. October 1998. doi:10.1046/j.1365-2958.1998.01062.x. PMID 9791175.
- ↑ "Signals, regulatory networks, and materials that build and break bacterial biofilms". Microbiology and Molecular Biology Reviews 73 (2): 310–47. June 2009. doi:10.1128/MMBR.00041-08. PMID 19487730.
- ↑ "Aminoglycoside antibiotics induce bacterial biofilm formation". Nature 436 (7054): 1171–5. August 2005. doi:10.1038/nature03912. PMID 16121184. Bibcode: 2005Natur.436.1171H. (primary source)
- ↑ "The promise and peril of transcriptional profiling in biofilm communities". Current Opinion in Microbiology 10 (3): 292–6. June 2007. doi:10.1016/j.mib.2007.05.011. PMID 17573234.
- ↑ 14.0 14.1 "Division of Labor: How Microbes Split Their Responsibility". Current Biology 28 (12): R697–R699. June 2018. doi:10.1016/j.cub.2018.05.024. PMID 29920261.
- ↑ Microbiology An Introduction (tenth ed.).
- ↑ "Determination of the van der Waals, electron donor and electron acceptor surface tension components of static Gram-positive microbial biofilms". Colloids Surf B Biointerfaces 21 (4): 299–310. August 2001. doi:10.1016/S0927-7765(00)00213-7. PMID 11397632.
- ↑ "Cellular hydrophobicity of Listeria monocytogenes involves initial attachment and biofilm formation on the surface of polyvinyl chloride". Lett. Appl. Microbiol. 50 (6): 618–25. June 2010. doi:10.1111/j.1472-765X.2010.02842.x. PMID 20438621.
- ↑ "7: Archaea" (in en). 6 February 2018. https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Bruslind)/07%3A_Archaea.
- ↑ Brock biology of microorganisms (Fifteenth, Global ed.). Pearson. 2019. p. 86. ISBN 9781292235103.
- ↑ 20.0 20.1 "Archaeal bundling pili of Pyrobaculum calidifontis reveal similarities between archaeal and bacterial biofilms". Proceedings of the National Academy of Sciences of the United States of America 119 (26): e2207037119. June 2022. doi:10.1073/pnas.2207037119. PMID 35727984. Bibcode: 2022PNAS..11907037W.
- ↑ 21.0 21.1 21.2 "Biofilms: Microbial Life on Surfaces". Emerging Infectious Diseases 8 (9): 881–890. 2002. doi:10.3201/eid0809.020063. PMID 12194761.
- ↑ "Biofilm matrix cloaks bacterial quorum sensing chemoattractants from predator detection". The ISME Journal 16 (5): 1388–1396. January 2022. doi:10.1038/s41396-022-01190-2. PMID 35034106.
- ↑ "Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics". Frontiers in Microbiology 10: 913. 2019. doi:10.3389/fmicb.2019.00913. PMID 31130925.
- ↑ "Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa". Journal of Bacteriology 189 (14): 5383–6. July 2007. doi:10.1128/JB.00137-07. PMID 17496081.
- ↑ 25.0 25.1 "Development of Antimicrobial Phototreatment Tolerance: Why the Methodology Matters". International Journal of Molecular Sciences (MDPI AG) 22 (4): 2224. February 2021. doi:10.3390/ijms22042224. PMID 33672375. 50px Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ "Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria". FEMS Microbiology Reviews (Oxford University Press (OUP)) 41 (3): 276–301. May 2017. doi:10.1093/femsre/fux010. PMID 28369412.
- ↑ "Biofilm formation as microbial development". Annual Review of Microbiology 54: 49–79. 2000. doi:10.1146/annurev.micro.54.1.49. PMID 11018124.
- ↑ 28.0 28.1 "Looking for chinks in the armor of bacterial biofilms". PLOS Biology 5 (11): e307. November 2007. doi:10.1371/journal.pbio.0050307. PMID 18001153.
- ↑ "Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity". Journal of Bacteriology 185 (16): 4693–8. August 2003. doi:10.1128/JB.185.16.4693-4698.2003. PMID 12896987.
- ↑ "Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms". Applied and Environmental Microbiology 74 (2): 470–6. January 2008. doi:10.1128/AEM.02073-07. PMID 18039822. Bibcode: 2008ApEnM..74..470I.
- ↑ "Enzymatic detachment of Staphylococcus epidermidis biofilms". Antimicrobial Agents and Chemotherapy 48 (7): 2633–6. July 2004. doi:10.1128/AAC.48.7.2633-2636.2004. PMID 15215120.
- ↑ "Biofilm-control strategies based on enzymic disruption of the extracellular polymeric substance matrix--a modelling study". Microbiology 151 (Pt 12): 3817–32. December 2005. doi:10.1099/mic.0.28165-0. PMID 16339929.
- ↑ "A fatty acid messenger is responsible for inducing dispersion in microbial biofilms". Journal of Bacteriology 191 (5): 1393–403. March 2009. doi:10.1128/JB.01214-08. PMID 19074399.
- ↑ "Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa". Journal of Bacteriology 188 (21): 7344–7353. 2006. doi:10.1128/jb.00779-06. PMID 17050922.
- ↑ "Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms". Microbial Biotechnology 2 (3): 370–378. 2009. doi:10.1111/j.1751-7915.2009.00098.x. PMID 21261931.
- ↑ "Dispersal of Biofilm in Cystic Fibrosis using Low Dose Nitric Oxide". University of Southampton. http://www.southampton.ac.uk/biosci/research/projects/dispersal_of_biofilms_in_cystic_fibrosis.page.
- ↑ 37.0 37.1 "Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles". Nature Communications 5: 4462. 2014. doi:10.1038/ncomms5462. PMID 25042103. Bibcode: 2014NatCo...5.4462C.
- ↑ "In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation". Nat Protoc 10 (8): 1165–80. August 2015. doi:10.1038/nprot.2015.067. PMID 26158442.
- ↑ 39.0 39.1 "Distinct bacterial population dynamics and disease dissemination after biofilm dispersal and disassembly". The ISME Journal 17 (8): 1290–1302. August 2023. doi:10.1038/s41396-023-01446-5. PMID 37270584.
- ↑ "The sociobiology of biofilms". FEMS Microbiology Reviews 33 (1): 206–24. January 2009. doi:10.1111/j.1574-6976.2008.00150.x. PMID 19067751.
- ↑ 41.0 41.1 41.2 41.3 "Pseudomonas aeruginosa Biofilm Infections: Community Structure, Antimicrobial Tolerance and Immune Response". Journal of Molecular Biology 427 (23): 3628–45. November 2015. doi:10.1016/j.jmb.2015.08.016. PMID 26319792.
- ↑ "Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture". Journal of Bacteriology 182 (12): 3593–6. June 2000. doi:10.1128/jb.182.12.3593-3596.2000. PMID 10852895.
- ↑ "A major protein component of the Bacillus subtilis biofilm matrix". Molecular Microbiology 59 (4): 1229–38. February 2006. doi:10.1111/j.1365-2958.2005.05020.x. PMID 16430696.
- ↑ "Salmonella biofilms using luminescent oligothiophenes". npj Biofilms and Microbiomes 2: 16024. 23 November 2016. doi:10.1038/npjbiofilms.2016.24. PMID 28721253.
- ↑ "Biofilms: an emergent form of bacterial life". Nature Reviews. Microbiology 14 (9): 563–75. August 2016. doi:10.1038/nrmicro.2016.94. PMID 27510863.
- ↑ "Liquid flow in biofilm systems". Applied and Environmental Microbiology 60 (8): 2711–6. August 1994. doi:10.1128/aem.60.8.2711-2716.1994. PMID 16349345. Bibcode: 1994ApEnM..60.2711S.
- ↑ "Control of cell fate by the formation of an architecturally complex bacterial community". Genes & Development 22 (7): 945–53. April 2008. doi:10.1101/gad.1645008. PMID 18381896.
- ↑ "Antibiotic resistance of bacteria in biofilms". Lancet 358 (9276): 135–8. July 2001. doi:10.1016/S0140-6736(01)05321-1. PMID 11463434. https://scholarworks.montana.edu/xmlui/handle/1/13570.
- ↑ "Characterisation of ESKAPE Pathogens with Special Reference to Multidrug Resistance and Biofilm Production in a Nepalese Hospital". Infect Drug Resist 14: 2201–2212. 2021. doi:10.2147/IDR.S306688. PMID 34163185.
- ↑ 50.0 50.1 50.2 50.3 50.4 50.5 "Bioelectric effect and bacterial biofilms. A systematic review". The International Journal of Artificial Organs 31 (9): 786–795. September 2008. doi:10.1177/039139880803100906. PMID 18924090.
- ↑ 51.0 51.1 "Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer". BMC Biology 12: 65. August 2014. doi:10.1186/s12915-014-0065-5. PMID 25124934.
- ↑ "Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure". Current Opinion in Biotechnology 14 (3): 255–61. June 2003. doi:10.1016/S0958-1669(03)00036-3. PMID 12849777.
- ↑ "Life after death: the critical role of extracellular DNA in microbial biofilms". Letters in Applied Microbiology 57 (6): 467–75. December 2013. doi:10.1111/lam.12134. PMID 23848166.
- ↑ "Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials". Journal of Bacteriology 183 (23): 6746–51. December 2001. doi:10.1128/JB.183.23.6746-6751.2001. PMID 11698361.
- ↑ "Introduction to Biofilms: Desirable and undesirable impacts of biofilm". http://www.cs.montana.edu/ross/personal/intro-biofilms-s3.htm. (primary source)
- ↑ "Influence of xylem fluid chemistry on planktonic growth, biofilm formation and aggregation of Xylella fastidiosa". FEMS Microbiology Letters 274 (2): 210–7. September 2007. doi:10.1111/j.1574-6968.2007.00827.x. PMID 17610515.
- ↑ "Biological wastewater treatment processes; secondary treatment". Staffordshire University. http://www.staffs.ac.uk/schools/sciences/consultancy/dladmin/zCIWEMWWT/Activity5/act5.html.
- ↑ Centre for Affordable Water and Sanitation Technology, Biosand Filter Manual: Design, Construction, & Installation," July 2007.
- ↑ "Slow Sand Filtration". Tech Brief (Morgantown, WV: National Drinking Water Clearinghouse (U.S.)) 14. June 2000. http://www.nesc.wvu.edu/pdf/DW/publications/ontap/tech_brief/TB15_SlowSand.pdf.
- ↑ "Plant Growth-Promoting Rhizobacteria on Canola (Rapeseed)". Plant Disease 72 (1): 42. 1988. doi:10.1094/pd-72-0042. ISSN 0191-2917.
- ↑ 61.0 61.1 61.2 61.3 "Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciens S499". FEMS Microbiology Ecology 79 (1): 176–91. January 2012. doi:10.1111/j.1574-6941.2011.01208.x. PMID 22029651. Bibcode: 2012FEMME..79..176N.
- ↑ "Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance (ISR)". Microbiological Research 164 (5): 493–513. September 2009. doi:10.1016/j.micres.2008.08.007. PMID 18845426.
- ↑ 63.0 63.1 "Plant responses to plant growth-promoting rhizobacteria". European Journal of Plant Pathology 119 (3): 243–254. 5 June 2007. doi:10.1007/s10658-007-9165-1. ISSN 0929-1873.
- ↑ 64.0 64.1 64.2 64.3 64.4 "Plant immune responses triggered by beneficial microbes". Current Opinion in Plant Biology 11 (4): 443–8. August 2008. doi:10.1016/j.pbi.2008.05.005. PMID 18585955.
- ↑ "Nitrogen-fixation by Azospirillum brasilense Cd is promoted when co-cultured with a mangrove rhizosphere bacterium (Staphylococcus sp.)". Soil Biology and Biochemistry 28 (12): 1651–1660. December 1996. doi:10.1016/s0038-0717(96)00251-9. ISSN 0038-0717.
- ↑ "Beneficial bacteria of agricultural importance". Biotechnology Letters 32 (11): 1559–70. November 2010. doi:10.1007/s10529-010-0347-0. PMID 20635120.
- ↑ "Induced Systemic Resistance by Fluorescent Pseudomonas spp". Phytopathology 97 (2): 239–43. February 2007. doi:10.1094/phyto-97-2-0239. PMID 18944381.
- ↑ "Induced Systemic Resistance Mediated by Plant Growth-Promoting Rhizobacteria (PGPR) and Fungi (PGPF)". Multigenic and Induced Systemic Resistance in Plants. Springer US. 2006. pp. 225–258. doi:10.1007/0-387-23266-4_10. ISBN 9780387232652.
- ↑ "Rhizosphere", eLS (John Wiley & Sons, Ltd), 21 August 2001, doi:10.1038/npg.els.0000403, ISBN 0470016175
- ↑ "Biofilms in the large bowel suggest an apparent function of the human vermiform appendix". Journal of Theoretical Biology 249 (4): 826–31. December 2007. doi:10.1016/j.jtbi.2007.08.032. PMID 17936308. Bibcode: 2007JThBi.249..826R.
- ↑ "Pathobiont release from dysbiotic gut microbiota biofilms in intestinal inflammatory diseases: a role for iron?". Journal of Biomedical Science 26 (1): 1. January 2019. doi:10.1186/s12929-018-0495-4. PMID 30602371.
- ↑ "Influence of Fouling Biofilms on Heat Transfer". Heat Transfer Engineering 3 (1): 23–37. 1981. doi:10.1080/01457638108939572. Bibcode: 1981HTrEn...3...23C. https://scholarworks.montana.edu/xmlui/handle/1/13866.
- ↑ "Impact of nitrate on the structure and function of bacterial biofilm communities in pipelines used for injection of seawater into oil fields". Applied and Environmental Microbiology 74 (9): 2841–51. May 2008. doi:10.1128/AEM.02027-07. PMID 18344353. Bibcode: 2008ApEnM..74.2841S.
- ↑ "Biofilms: A microbial home". Journal of Indian Society of Periodontology 15 (2): 111–4. April 2011. doi:10.4103/0972-124X.84377. PMID 21976832.
- ↑ "Microbial biofilms in dental medicine in reference to implanto-prostethic rehabilitation" (in ro). Revista de Chirurgie Oro-maxilo-facială și Implantologie 1 (1): 9–13. December 2010. 8. ISSN 2069-3850. http://www.revistaomf.ro/(8). Retrieved 3 June 2012.[yes|permanent dead link|dead link}}](webpage has a translation button)
- ↑ "Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms". Journal of Industrial Microbiology 15 (3): 198–207. September 1995. doi:10.1007/bf01569826. PMID 8519478.
- ↑ 77.0 77.1 77.2 "Responses of cariogenic streptococci to environmental stresses". Current Issues in Molecular Biology 7 (1): 95–107. January 2005. PMID 15580782. http://www.horizonpress.com/cimb/v/v7/07.pdf. Retrieved 3 April 2014.
- ↑ "The formation apurinic acid from the desoxyribonucleic acid of calf thymus". The Journal of Biological Chemistry 195 (1): 49–63. March 1952. doi:10.1016/S0021-9258(19)50874-2. PMID 14938354.
- ↑ "Transitions and transversions induced by depurinating agents". Proceedings of the National Academy of Sciences of the United States of America 47 (4): 540–5. April 1961. doi:10.1073/pnas.47.4.540. PMID 13701660. Bibcode: 1961PNAS...47..540B.
- ↑ 80.0 80.1 Pennwell, "Toothbrush technology, dentifrices and dental biofilm removal." Dental Academy of CE Accessed 12 January 2022
- ↑ Pathology of dental caries. In: Dental caries: the disease and its clinical management.. Oxford (UK): Wiley Blackwell. 2015. pp. 7–9. ISBN 978-1405138895.
- ↑ 82.0 82.1 "Natural genetic transformation of Streptococcus mutans growing in biofilms". J. Bacteriol. 183 (3): 897–908. February 2001. doi:10.1128/JB.183.3.897-908.2001. PMID 11208787.
- ↑ "Quorum Sensing and Biofilm Formation by Streptococcus mutans". Bacterial Signal Transduction: Networks and Drug Targets. Advances in Experimental Medicine and Biology. 631. 2008. pp. 178–88. doi:10.1007/978-0-387-78885-2_12. ISBN 978-0-387-78884-5. https://archive.org/details/bacterialsignalt0000unse/page/178.
- ↑ 84.0 84.1 "Adaptive value of sex in microbial pathogens". Infect. Genet. Evol. 8 (3): 267–85. May 2008. doi:10.1016/j.meegid.2008.01.002. PMID 18295550.http://www.hummingbirds.arizona.edu/Faculty/Michod/Downloads/IGE%20review%20sex.pdf
- ↑ "Biofilm development on Caenorhabditis elegans by Yersinia is facilitated by quorum sensing-dependent repression of type III secretion". PLOS Pathogens 7 (1): e1001250. January 2011. doi:10.1371/journal.ppat.1001250. PMID 21253572.
- ↑ "Biofilm matrix disrupts nematode motility and predatory behavior". The ISME Journal 15 (1): 260–269. September 2020. doi:10.1038/s41396-020-00779-9. PMID 32958848.
- ↑ "Biofilm matrix cloaks bacterial quorum sensing chemoattractants from predator detection". The ISME Journal 16 (5): 1388–1396. May 2022. doi:10.1038/s41396-022-01190-2. PMID 35034106.
- ↑ "Biofilm formation and dispersal in Gram-positive bacteria". Current Opinion in Biotechnology 22 (2): 172–9. April 2011. doi:10.1016/j.copbio.2010.10.016. PMID 21109420. https://pure.rug.nl/ws/files/6762350/2011CurrOpinBiotechnolAbee2.pdf.
- ↑ "Role of cyanobacterial exopolysaccharides in phototrophic biofilms and in complex microbial mats". Life 5 (2): 1218–38. April 2015. doi:10.3390/life5021218. PMID 25837843. Bibcode: 2015Life....5.1218R.
- ↑ 90.0 90.1 "Biofilm formation by plant-associated bacteria". Annual Review of Microbiology 61: 401–22. 2007. doi:10.1146/annurev.micro.61.080706.093316. PMID 17506679.
- ↑ 91.0 91.1 "Microbial exopolymers link predator and prey in a model yeast biofilm system". Microb. Ecol. 52 (2): 187–97. August 2006. doi:10.1007/s00248-006-9063-7. PMID 16897306.
- ↑ 92.0 92.1 "Ecology of intertidal microbial biofilms: Mechanisms, patterns and future research needs". Journal of Sea Research 92: 2–5. 2014. doi:10.1016/j.seares.2014.07.003. Bibcode: 2014JSR....92....2V.
- ↑ 93.0 93.1 "Production and Characterization of the Intra- and Extracellular Carbohydrates and Polymeric Substances (EPS) of Three Sea-Ice Diatom Species, and Evidence for a Cryoprotective Role for EPS". J. Phycol. 48 (6): 1494–509. December 2012. doi:10.1111/jpy.12004. PMID 27009999.
- ↑ "Research on microbial biofilms (PA-03-047)". NIH, National Heart, Lung, and Blood Institute. 20 December 2002. http://grants.nih.gov/grants/guide/pa-files/PA-03-047.html.
- ↑ Molecular Oral Microbiology. Caister Academic Press. 2008. pp. 88–91. ISBN 978-1-904455-24-0.
- ↑ "Fusarium and Candida albicans biofilms on soft contact lenses: model development, influence of lens type, and susceptibility to lens care solutions". Antimicrobial Agents and Chemotherapy 52 (1): 171–82. January 2008. doi:10.1128/AAC.00387-07. PMID 17999966.
- ↑ "Propionibacterium acnes biofilm is present in intervertebral discs of patients undergoing microdiscectomy". PLOS ONE 12 (4): e0174518. 3 April 2017. doi:10.1371/journal.pone.0174518. PMID 28369127. Bibcode: 2017PLoSO..1274518C.
- ↑ "Riddle of biofilm resistance". Antimicrobial Agents and Chemotherapy 45 (4): 999–1007. April 2001. doi:10.1128/AAC.45.4.999-1007.2001. PMID 11257008.
- ↑ "Bacterial biofilms: an emerging link to disease pathogenesis". Annual Review of Microbiology 57: 677–701. 2003. doi:10.1146/annurev.micro.57.030502.090720. PMID 14527295.
- ↑ 100.0 100.1 "High Prevalence of Biofilms on Retrieved Implants from Aseptic Pseudarthrosis Cases". Spine Surgery and Related Research 5 (2): 104–108. 2020. doi:10.22603/ssrr.2020-0147. PMID 33842718.
- ↑ 101.0 101.1 "New study first to visually capture biofilm architecture in retrieved implants from live patients" (in en-GB). 20 November 2020. https://spinalnewsinternational.com/new-study-first-to-visually-capture-biofilm-architecture-in-retrieved-implants-from-live-patients/.
- ↑ "biofilm". 22 December 2020. https://orthospinenews.com/2020/12/22/first-study-to-visually-capture-biofilm-structure-of-retrieved-implants-from-spine-patients-with-pseudarthrosis/.
- ↑ "Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo". Wound Repair and Regeneration 16 (1): 23–9. 2008. doi:10.1111/j.1524-475X.2007.00303.x. PMID 18211576.
- ↑ "Detection of Biofilm in Wounds as an Early Indicator for Risk for Tissue Infection and Wound Chronicity". Annals of Plastic Surgery 76 (1): 127–31. January 2016. doi:10.1097/SAP.0000000000000440. PMID 25774966.
- ↑ "Bacterial biofilms in surgical specimens of patients with chronic rhinosinusitis". The Laryngoscope 115 (4): 578–82. 2005. doi:10.1097/01.mlg.0000161346.30752.18. PMID 15805862.
- ↑ "Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis". The Laryngoscope 116 (7): 1121–6. July 2006. doi:10.1097/01.mlg.0000221954.05467.54. PMID 16826045.
- ↑ "Optical imaging of bacterial infection in living mice using a fluorescent near-infrared molecular probe". Journal of the American Chemical Society 128 (51): 16476–7. December 2006. doi:10.1021/ja0665592. PMID 17177377.
- ↑ "Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus". mBio 3 (4): e00198-12. 2012. doi:10.1128/mBio.00198-12. PMID 22851659.
- ↑ The tragedy of the commons and prisoner's dilemma may improve our realization of the theory of life and provide us with advanced therapeutic ways (Report). 2015. doi:10.13140/RG.2.1.2327.9842.
- ↑ "Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients". Advanced Drug Delivery Reviews 85: 7–23. May 2015. doi:10.1016/j.addr.2014.11.017. PMID 25477303.
- ↑ "Extracellular DNA required for bacterial biofilm formation". Science 295 (5559): 1487. February 2002. doi:10.1126/science.295.5559.1487. PMID 11859186.
- ↑ "Human skin wounds: a major and snowballing threat to public health and the economy". Wound Repair and Regeneration 17 (6): 763–771. November 2009. doi:10.1111/j.1524-475X.2009.00543.x. PMID 19903300.
- ↑ "A component of innate immunity prevents bacterial biofilm development". Nature 417 (6888): 552–555. May 2002. doi:10.1038/417552a. PMID 12037568. Bibcode: 2002Natur.417..552S.
- ↑ "Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis". Molecular Microbiology 61 (5): 1196–210. September 2006. doi:10.1111/j.1365-2958.2006.05310.x. PMID 16925554.
- ↑ "Fratricide is essential for efficient gene transfer between pneumococci in biofilms". Appl. Environ. Microbiol. 78 (16): 5897–905. August 2012. doi:10.1128/AEM.01343-12. PMID 22706053. Bibcode: 2012ApEnM..78.5897W.
- ↑ "The insect antimicrobial peptide cecropin A disrupts uropathogenic Escherichia coli biofilms". npj Biofilms and Microbiomes 6 (1): 6. 2020. doi:10.1038/s41522-020-0116-3. PMID 32051417.
- ↑ "The role of quorum sensing in Escherichia coli (ETEC) virulence factors". Veterinary Microbiology 180 (3–4): 245–252. November 2015. doi:10.1016/j.vetmic.2015.08.015. PMID 26386492.
- ↑ "Life on the outside: role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli". Frontiers in Microbiology 5: 317. 2014. doi:10.3389/fmicb.2014.00317. PMID 25071733.
- ↑ "Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture". Journal of Bacteriology 182 (12): 3593–3596. June 2000. doi:10.1128/JB.182.12.3593-3596.2000. PMID 10852895.
- ↑ "Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients". The Indian Journal of Medical Research 139 (6): 945–948. June 2014. PMID 25109731.
- ↑ "Type 1 fimbriae contribute to catheter-associated urinary tract infections caused by Escherichia coli". Journal of Bacteriology 196 (5): 931–939. March 2014. doi:10.1128/JB.00985-13. PMID 24336940.
- ↑ "Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection". The Journal of Infectious Diseases 204 (6): 937–941. September 2011. doi:10.1093/infdis/jir441. PMID 21849291.
- ↑ "Toxin-induced necroptosis is a major mechanism of Staphylococcus aureus lung damage". PLOS Pathogens 11 (4): e1004820. April 2015. doi:10.1371/journal.ppat.1004820. PMID 25880560.
- ↑ "Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo". Journal of Immunology 186 (11): 6585–6596. June 2011. doi:10.4049/jimmunol.1002794. PMID 21525381.
- ↑ "Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype". MedChemComm 10 (8): 1231–1241. August 2019. doi:10.1039/c9md00044e. PMID 31534648.
- ↑ "Quorum sensing in biofilms—how to destroy the bacterial citadels or their cohesion/power?". Anaerobe 17 (6): 280–5. December 2011. doi:10.1016/j.anaerobe.2011.03.023. PMID 21497662.
- ↑ Biofilm infections. New York: Springer. 2011. ISBN 9781441960832. OCLC 682907381.
- ↑ 128.0 128.1 128.2 128.3 "Medical biofilms". Biotechnology and Bioengineering 100 (1): 1–18. May 2008. doi:10.1002/bit.21838. PMID 18366134.
- ↑ "Biofilm formation on intrauterine devices in patients with recurrent vulvovaginal candidiasis". Medical Mycology 48 (1): 211–6. February 2010. doi:10.3109/13693780902856626. PMID 20055746.
- ↑ "Antibiotic Resistance Related to Biofilm Formation in Klebsiella pneumoniae". Pathogens 3 (3): 743–758. September 2014. doi:10.3390/pathogens3030743. PMID 25438022.
- ↑ "Evolutionary pathways to antibiotic resistance are dependent upon environmental structure and bacterial lifestyle". eLife 8: e47612. September 2019. doi:10.7554/eLife.47612. PMID 31516122.
- ↑ "A growing battlefield in the war against biofilm-induced antimicrobial resistance: insights from reviews on antibiotic resistance". Front Cell Infect Microbiol 13: 1327069. 2023. doi:10.3389/fcimb.2023.1327069. PMID 38188636.
- ↑ 133.0 133.1 "Effect of electrical energy on the efficacy of biofilm treatment using the bioelectric effect". npj Biofilms and Microbiomes 1: 15016. 2015. doi:10.1038/npjbiofilms.2015.16. PMID 28721233.
- ↑ "Genomic Insights into Oil Biodegradation in Marine Systems". Microbial Biodegradation: Genomics and Molecular Biology. Horizon Scientific Press. 2008. pp. 1971. ISBN 978-1-904455-17-2. https://books.google.com/books?id=wx6v3TIkzXUC&pg=PA1971.
- ↑ "A stable synergistic microbial consortium for simultaneous azo dye removal and bioelectricity generation". Bioresource Technology 155: 71–76. March 2014. doi:10.1016/j.biortech.2013.12.078. PMID 24434696.
- ↑ "Engineering PQS biosynthesis pathway for enhancement of bioelectricity production in pseudomonas aeruginosa microbial fuel cells". PLOS ONE 8 (5): e63129. 2013. doi:10.1371/journal.pone.0063129. PMID 23700414. Bibcode: 2013PLoSO...863129W.
- ↑ "Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation—part A". Appl. Microbiol. Biotechnol. 97 (17): 7529–41. September 2013. doi:10.1007/s00253-013-4954-2. PMID 23720034.
- ↑ "Microbial–Enzymatic Combinatorial Approach to Capture and Release Microplastics" (in en). Environmental Science & Technology Letters 9 (11): 975–982. 2022-10-12. doi:10.1021/acs.estlett.2c00558. ISSN 2328-8930. Bibcode: 2022EnSTL...9..975C.
- ↑ "Engineering a microbial 'trap and release' mechanism for microplastics removal" (in en). Chemical Engineering Journal 404: 127079. 2021-01-15. doi:10.1016/j.cej.2020.127079. ISSN 1385-8947.
- ↑ 140.0 140.1 140.2 140.3 140.4 140.5 140.6 140.7 140.8 "Biofilm formation in food industries: A food safety concern". Food Control 31 (2): 572–585. June 2013. doi:10.1016/j.foodcont.2012.12.001. ISSN 0956-7135.
- ↑ T. Tarver, "Biofilms: A Threat to Food Safety – IFT.org", Ift.org, 2016.
- ↑ 142.0 142.1 142.2 142.3 "Significance of microbial biofilms in food industry: a review". International Journal of Food Microbiology 42 (1–2): 9–27. June 1998. doi:10.1016/s0168-1605(98)00060-9. PMID 9706794.
- ↑ "Biofilm dispersal induced by mechanical cutting leads to heightened foodborne pathogen dissemination". Food Microbiology 102: 103914. April 2022. doi:10.1016/j.fm.2021.103914. PMID 34809940.
- ↑ 144.0 144.1 144.2 144.3 "Microbial biofilms in seafood: A food-hygiene challenge". Food Microbiology 49: 41–55. 2015. doi:10.1016/j.fm.2015.01.009. PMID 25846914.
- ↑ "Rhamnolipid and surfactin inhibit Listeria monocytogenes adhesion". Food Research International 44 (1): 481–488. January 2011. doi:10.1016/j.foodres.2010.09.002.
- ↑ "Effect of berberine on Staphylococcus epidermidis biofilm formation". International Journal of Antimicrobial Agents 34 (1): 60–6. July 2009. doi:10.1016/j.ijantimicag.2008.10.033. PMID 19157797.
- ↑ "Fire Ant Venom Alkaloids Inhibit Biofilm Formation". Toxins 11 (7): 420. July 2019. doi:10.3390/toxins11070420. PMID 31323790.
- ↑ "Marine biofouling on fish farms and its remediation". Advances in Marine Biology 47: 215–252. 2004. doi:10.1016/S0065-2881(04)47003-5. ISBN 9780120261482. PMID 15596168.
- ↑ "Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture". Marine Biotechnology 9 (4): 399–410. 2007. doi:10.1007/s10126-007-9001-9. PMID 17497196.
- ↑ "Biofilm formation by the fish pathogen Flavobacterium columnare: development and parameters affecting surface attachment". Applied and Environmental Microbiology 79 (18): 5633–42. September 2013. doi:10.1128/AEM.01192-13. PMID 23851087. Bibcode: 2013ApEnM..79.5633C.
- ↑ "Identification of bacterial pathogens in biofilms of recirculating aquaculture systems". Journal of Aquatic Food Product Technology 13: 125–133. 2004. doi:10.1300/j030v13n01_11.
- ↑ "Biofilm development within a larval rearing tank of the tropical rock lobster, Panulirus ornatus". Aquaculture 260 (1–4): 27–38. 2006. doi:10.1016/j.aquaculture.2006.06.023.
- ↑ "Effects of seawater ozonation on biofilm development in aquaculture tanks". Systematic and Applied Microbiology 32 (4): 266–77. July 2009. doi:10.1016/j.syapm.2009.04.001. PMID 19446976.
- ↑ "Rapid detection of microorganisms in a fish infection microfluidics platform". Journal of Hazardous Materials 431: 128572. June 2022. doi:10.1016/j.jhazmat.2022.128572. PMID 35278965.
- ↑ "Mass mortality of Penaeus monodon larvae due to antibiotic-resistant Vibrio harveyi infection". Aquaculture 128 (3–4): 203–209. 1994. doi:10.1016/0044-8486(94)90309-3.
- ↑ "Optical sectioning of microbial biofilms". Journal of Bacteriology 173 (20): 6558–67. October 1991. doi:10.1128/jb.173.20.6558-6567.1991. PMID 1917879.
- ↑ "Adhesion of bacteria and diatoms to surfaces in the sea: a review". Aquatic Microbial Ecology 9 (1): 87–96. 1995. doi:10.3354/ame009087.
- ↑ "Fungal Biofilms". PLOS Pathog 8 (4): e1002585. 2012. doi:10.1371/journal.ppat.1002585. PMID 22496639.
- ↑ "Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance". J. Bacteriol. 183 (18): 5385–94. September 2001. doi:10.1128/jb.183.18.5385-5394.2001. PMID 11514524.
- ↑ Microbial Biofilms: Current Research and Applications. Horizon Scientific Press. 2012. pp. 61–71. ISBN 978-1904455967.
- ↑ "Protection of cells from salinity stress by extracellular polymeric substances in diatom biofilms". Biofouling 30 (8): 987–98. September 2014. doi:10.1080/08927014.2014.960859. PMID 25268215.
- ↑ "Biofilm and capsule formation of the diatom Achnanthidium minutissimum are affected by a bacterium". J. Phycol. 51 (2): 343–55. April 2015. doi:10.1111/jpy.12280. PMID 26986529. Bibcode: 2015JPcgy..51..343W. http://nbn-resolving.de/urn:nbn:de:bsz:352-0-289776.
- ↑ "Photoautotrophic-heterotrophic biofilm communities: a laboratory incubator designed for growing axenic diatoms and bacteria in defined mixed-species biofilms". Environ Microbiol Rep 4 (1): 133–40. February 2012. doi:10.1111/j.1758-2229.2011.00315.x. PMID 23757240. http://nbn-resolving.de/urn:nbn:de:bsz:352-177088.
- ↑ "Mechanisms of, and barriers to, horizontal gene transfer between bacteria". Nature Reviews. Microbiology 3 (9): 711–721. September 2005. doi:10.1038/nrmicro1234. PMID 16138099.
- ↑ 165.0 165.1 165.2 165.3 165.4 165.5 165.6 165.7 165.8 "Formation, Development, and Cross-Species Interactions in Biofilms". Frontiers in Microbiology 12: 757327. 2022. doi:10.3389/fmicb.2021.757327. PMID 35058893.
- ↑ "The F-pilus biomechanical adaptability accelerates conjugative dissemination of antimicrobial resistance and biofilm formation". Nature Communications 14 (1): 1879. April 2023. doi:10.1038/s41467-023-37600-y. PMID 37019921.
- ↑ "Suicide and fratricide in bacterial biofilms". The International Journal of Artificial Organs 32 (9): 537–544. September 2009. doi:10.1177/039139880903200902. PMID 19851979.
- ↑ 168.00 168.01 168.02 168.03 168.04 168.05 168.06 168.07 168.08 168.09 "Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism". FEMS Microbiology Ecology 96 (5). May 2020. doi:10.1093/femsec/fiaa031. PMID 32109282.
- ↑ "Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer". BMC Biology 12 (1): 65. August 2014. doi:10.1186/s12915-014-0065-5. PMID 25124934.
- ↑ "Critical review on biofilm methods". Critical Reviews in Microbiology 43 (3): 313–351. May 2017. doi:10.1080/1040841X.2016.1208146. PMID 27868469.
Further reading
- Community structure and co-operation in biofilms. Cambridge, UK: Cambridge University Press. 2000. ISBN 978-0-521-79302-5.
- Microbial biofilms. Cambridge, UK: Cambridge University Press. 2003. ISBN 978-0-521-54212-8.
- Biofilms in the food and beverage industries. Woodhead Publishing Limited. 2009. ISBN 978-1-84569-477-7.
External links
- A TED-ED animation on basic biofilm biology: The microbial jungles all over the place (and you) by Scott Chimileski and Roberto Kolter
- Thickness analysis, organic and mineral proportion of biofilms in order to decide a treatment strategy
- Biofilm Archive of Biofilm Research & News
- "Why Am I Still Sick?" – The Movie, 2012: Documentary on Biofilms: The Silent Role of Biofilms in Chronic Disease
- HD Video Interviews on biofilms, antibiotics, etc. with experts, youtube.com: ADRSupport/biofilm
 |