Biology:Cowpea chlorotic mottle virus
| Cowpea chlorotic mottle virus | |
|---|---|
| |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Kitrinoviricota |
| Class: | Alsuviricetes |
| Order: | Martellivirales |
| Family: | Bromoviridae |
| Genus: | Bromovirus |
| Species: | Cowpea chlorotic mottle virus
|
Cowpea chlorotic mottle virus, known by the abbreviation CCMV, is a virus that specifically infects the cowpea plant, or black-eyed pea. The leaves of infected plants develop yellow spots, hence the name "chlorotic". Similar to its "brother" virus, Cowpea mosaic virus (CPMV), CCMV is produced in high yield in plants. In the natural host, viral particles can be produced at 1–2 mg per gram of infected leaf tissue. Belonging to the bromovirus genus, cowpea chlorotic mottle virus (CCMV) is a small spherical plant virus. Other members of this genus include the brome mosaic virus (BMV) and the broad bean mottle virus (BBMV).
History
Bancroft et al. in 1967 described the first experiments to isolate and characterize the virus. Since that time, due to the relative ease with which it is grown and isolated, many researchers have focused their attention on the virus. The interest of the scientific community for this virus is also due to a conspicuous property: it is possible to disassemble the virus and remove the genetic material, the RNA. Then, under slightly acidic pH and with relatively high amounts of salts, it is possible to stimulate the self-assembly of the protein subunits, into a shell of identical size to the virus. This yields an empty capsid which has a number of interesting properties. Several successful attempts are reported to incorporate other materials, such as inorganic crystals, inside the capsid. This could lead to possible drug treatments in the future.[citation needed]
Genome and structure
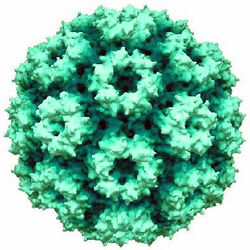
CCMV is composed of an icosahedral protein capsid (T=3)[1] that is 28 nm in diameter. This capsid is constructed by 180 identical protein subunits each with a primary structure of 190 amino acid residues. There are three subunits are distributed over the virus coat, A, B, and C. The A subunits are arranged in pentamers and the B and C subunits are together arranged in hexamers. The virus coat is built up from 12 pentamers and 20 hexamers. Inside the capsid lies the (+)ssRNA genome consisting of around 3000 nucleotides.[2] The genome is divided into three parts (RNA-1-3) with a subgenomic portion referred to as RNA4.[1] RNA-1, with a heavy density, is surrounded by its own capsid. RNA-2, with a light density, also has its own capsid. Because RNA-3 and RNA-4 are medium-density, they are encapsidated together. RNA-1 and RNA-2 are thought to be involved in viral replication while RNA-3 has a role in the spread of infection throughout the plant.[3] When RNA-3 is deficient, virus replication still does occur, just at a significantly reduced level. Due to these four species of single-stranded, positive sense RNA molecules, the CCMV genome codes for four separate genes.[2]
Lipofectamine is a reagent used in the lab to aid in transfection, allowing foreign DNA to enter the target cell. In a study by Garmann et al. they found that the CCMV viral capsids are very robust, remaining intact even after treatment with RNase in the absence of lipofectamine.[2]
Entry into host cell and interaction
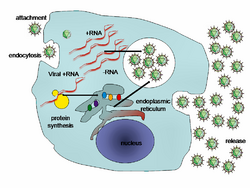
There is not a lot known about plant virus and host cell interaction due to the difficulty of studying organisms with cell walls. One study examined the interactions between CCMV and cowpea protoplasts and found that it was dependent on aspecific binding, mostly relying on electrostatic interactions between the plasma membrane and virus particles, specifically negatively charged vesicles and the positively charged N-terminal arm of viral coat proteins, further labelling CCMV as an endocytic virus. It also takes advantage of membrane lesions to introduce viral particles into the cell. Overall, the most effective infection occurred by internalization through membrane lesions of the host.[4]
One specific protein, ORF3a is a movement protein present in the CCMV genome which helps transport the viral genome to neighbouring plant cells using the plasmodesmata. This allows the virus to bypass the host cell wall barrier and effectively infect the host. The movement of CCMV requires no budding because the tubule structures enlarge the plasmodesmata enough to allow the direct passage of the viral capsid through the cell wall.[5]
Typical virus infection involves an exponential increase in virus concentration followed by a rapid decline of virus replication. In the presence of RNA 3 deficiency, virus replication still does occur, just at a significantly reduced level. It also is thought to be responsible for a low coat protein to viral RNA ratio.[citation needed]
Replication cycle
After virus entry, the protein capsid is degraded by the host cell, and this allows the unpackaging of the viral RNA. RNA1 and RNA2 encode for protein 1a and 2a-polymerase, respectively, both of which are expressed to produce viral replication proteins within the cell.[6] The actual replication process occurs in membrane vesicles created from invaginations of the host endoplasmic reticulum membrane. The viral RNA is replicated into a dsRNA genome utilizing an RNA dependent-RNA polymerase. The newly synthesized dsRNA is used to both transcribe more (+)ssRNA from the template (-)RNA strand and the existing (+)RNA strand is replicated to produce many copies to use as translatable mRNA. During this process, subgenomic RNA4 is also translated to produce viral capsid proteins. Using the newly synthesized copies of (+)ssRNA and capsid proteins, the virus assembles within the vesicle.[7]
Recombination
Upon joint infection of plant host cells with two different CCMV gene deletion mutants, functional RNA virus genomes can be regenerated by homologous recombination repair.[8] The mechanism of recombination is likely strand switching (copy choice) during viral RNA replication. The rapidity and frequency of this recombination suggests that such genome rescue is probably significant in natural populations of CCMV.[8]
Assembly and release
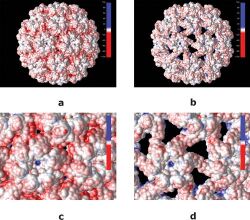
The assembly of a virus is key to its efficacy as it needs to be both stable enough to protect its genome before entry into the cell and labile enough to release its genetic contents into target cell as it disassembles. The single stranded RNA is threaded through small pores already present in the capsid. At a neutral pH, the capsid protein reversibly binds to RNA forming a pre-capsid complex. This consists of RNA surrounded by enough capsid proteins (CP) to neutralize the negative charges of the RNA phosphate backbone. When acidification occurs, an irreversible conformational change occurs making the final product of an icosahedral capsid. This is done by sending any excess CPs from the RNA to the outside of the new capsid. This process is reliant on the basicity of the CP due to its N-terminal arginine-rich motif (ARM) and the capsid exterior negative charge density. The capsid protein is also involved in viral movement, transmission, symptom expression, and targeted hosts.[9] As seen above, the assembly of CCMV is a pH dependent mechanism and so is the disassembly. At a pH of 5, CCMV is stable, but at a pH of 7.0 and without ions like Ca2+ or Mg2+ swelling of the capsid diameter occurs. This creates openings in the capsid, but the viral RNA is not released at this time, allowing this process to be reversed. This is important because calcium ions have been found to be essential to viral stability. Although RNA is not spontaneously released, when swelling occurs and the virus is in a proper environment for infection, the swelling will cause RNA release into the cytoplasm of a target cell.[10]
The figure on the right illustrates CCMV in acidic conditions (a) and CCMV as the pH changes and swelling occurs (b), this allows for electrostatic interactions, further enhancing the virus's ability to infect a host.[citation needed]
Symptomology
This virus has been observed to infect only plant cells, specifically Cowpeas. The primarily observed symptom of CCMV is bright chlorosis, or yellow coloring, in the leaves of the plant, known as the CCMV-T strain. This chlorosis has been observed as a less severe effect, producing a light green coloration when infecting plants with an attenuated strain, termed CCMV-M. Results from an experiment conducted by de Assis Filho et al. indicated that this primary symptom was caused by the amino acid at position 151 of the capsid coat protein.[11]
Vectors and transmission
CCMV has been found to be transmitted by the bean leaf beetle, Cerotoma trifurcata, and the spotted cucumber beetle, Diabrotica undecimpunctata howardii. CCMV affects beans and cowpeas, but it has been found that the viral replication is much greater when a virus is acquired from and transmitted to beans rather than cowpeas.[12]
As discussed in the “Assembly and Release” section, CCMV is stabilized by acidic conditions (pH = 5.0) it is thought that the intestines of insects provide the acidic conditions to allow CCMV's transmission and stability.[13]
Recent studies in yeast
In December 2018, CCMV replication was fully reconstituted in Saccharomyces cerevisiae, a type of yeast. In this experiment, it was found that protein 1a was the only viral factor needed to induce invagination of the endoplasmic reticulum and begin the replication process. The 2a polymerase was found to be recruited by protein 1a after the formation of the replication spherule. One limitation was realized for the replication of CCMV in S. cerevisiae, and this was due to the lack of RNA-3 replication. The significance of this experiment stretches beyond the scope of the results, because S. cerevisiae is a popular model organism for viral inoculation and may open avenues for further research with CCMV.[6]
Associated viruses
The following viruses are closely related to CCMV and are members of the Bromovirus genus:[14]
- Broad bean mottle virus
- Brome mosaic virus
- Cassia yellow blotch virus
- Melandrium yellow fletch virus
- Spring beauty latent virus
References
- ↑ 1.0 1.1 "Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy". Structure 3 (1): 63–78. January 1995. doi:10.1016/S0969-2126(01)00135-6. PMID 7743132.
- ↑ 2.0 2.1 2.2 Garmann RF (2014). The Self-assembly of Cowpea Chlorotic Mottle Virus. UCLA Electronic Theses and Dissertations (Thesis). UCLA. Retrieved 10 March 2019.
- ↑ Horst, R. Kenneth (2008). "Bean Yellow Stipple = Cowpea Chlorotic Mottle Bromovirus Yellow Stipple=Cowpea Chlorotic Mottle Bromovirus". Westcott's Plant Disease Handbook (Seventh ed.). Springer Netherlands. pp. 610. doi:10.1007/978-1-4020-4585-1_986. ISBN 9781402045844. https://archive.org/details/westcottsplantdi00hors.
- ↑ Roenhorst, Johanna (10 October 1989). Early Stages in Cowpea Chlorotic Mottle Virus Infection (Thesis). Retrieved 11 March 2019.
- ↑ "ORF3a - Movement protein - Cowpea chlorotic mottle virus (CCMV) - ORF3a gene & protein". https://www.uniprot.org/uniprot/P20180.
- ↑ 6.0 6.1 "Cowpea chlorotic mottle bromovirus replication proteins support template-selective RNA replication in Saccharomyces cerevisiae". PLOS ONE 13 (12): e0208743. 2018-12-26. doi:10.1371/journal.pone.0208743. PMID 30586378. Bibcode: 2018PLoSO..1308743S.
- ↑ "Bromoviridae ~ ViralZone page". https://viralzone.expasy.org/31?outline=all_by_species.
- ↑ 8.0 8.1 Allison R, Thompson C, Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1820-4. doi: 10.1073/pnas.87.5.1820. PMID 2308940; PMCID: PMC53575
- ↑ "The assembly pathway of an icosahedral single-stranded RNA virus depends on the strength of inter-subunit attractions". Journal of Molecular Biology 426 (5): 1050–60. March 2014. doi:10.1016/j.jmb.2013.10.017. PMID 24148696.
- ↑ "Electrostatic properties of cowpea chlorotic mottle virus and cucumber mosaic virus capsids". Biopolymers 82 (2): 106–20. June 2006. doi:10.1002/bip.20409. PMID 16278831.
- ↑ "Symptom induction by Cowpea chlorotic mottle virus on Vigna unguiculata is determined by amino acid residue 151 in the coat protein". The Journal of General Virology 83 (Pt 4): 879–83. April 2002. doi:10.1099/0022-1317-83-4-879. PMID 11907338.
- ↑ Hobbs, HA; Fulton, JB (September 11, 1978). "Beetle Transmission of Cowpea Chlorotic Mottle Virus". Phytopathology 69 (3): 255–256. doi:10.1094/Phyto-69-255. https://www.apsnet.org/publications/phytopathology/backissues/Documents/1979Articles/Phyto69n03_255.PDF.
- ↑ Wilts, Bodo D.; Schaap, Iwan A. T.; Schmidt, Christoph F. (May 2015). "Swelling and Softening of the Cowpea Chlorotic Mottle Virus in Response to pH Shifts". Biophysical Journal 108 (10): 2541–2549. doi:10.1016/j.bpj.2015.04.019. PMID 25992732. Bibcode: 2015BpJ...108.2541W.
- ↑ "International Committee on Taxonomy of Viruses (ICTV)" (in en). https://ictv.global/taxonomy.
Further reading
- Meijer, Sander; Feenstra, Anton. "CCMV Overview". Biochemistry Homepage. Wageningen University. http://gcg.tran.wau.nl/ccmv-overview/CCMV.html.
- "Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus". Virology 32 (2): 354–6. June 1967. doi:10.1016/0042-6822(67)90284-X. PMID 6025882.
- "Host–guest encapsulation of materials by assembled virus protein cages.". Nature 393 (6681): 152–155. May 1998. doi:10.1038/30211. Bibcode: 1998Natur.393..152D.
- "Folic acid-mediated targeting of cowpea mosaic virus particles to tumor cells". Chemistry & Biology 14 (10): 1152–62. October 2007. doi:10.1016/j.chembiol.2007.08.015. PMID 17961827.
- "Utilisation of plant viruses in bionanotechnology". Organic & Biomolecular Chemistry 5 (18): 2891–902. September 2007. doi:10.1039/b708175h. PMID 17728853.
External links
- ICTVdB - The Universal Virus Database: Cowpea chlorotic mottle virus
- Family Groups - The Baltimore Method
Wikidata ☰ Q5179790 entry
 |