Biology:Tiling array
Tiling arrays are a subtype of microarray chips. Like traditional microarrays, they function by hybridizing labeled DNA or RNA target molecules to probes fixed onto a solid surface.
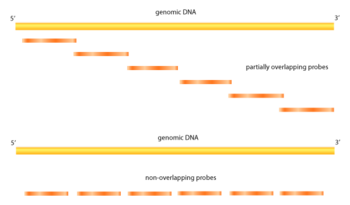
Tiling arrays differ from traditional microarrays in the nature of the probes. Instead of probing for sequences of known or predicted genes that may be dispersed throughout the genome, tiling arrays probe intensively for sequences which are known to exist in a contiguous region. This is useful for characterizing regions that are sequenced, but whose local functions are largely unknown. Tiling arrays aid in transcriptome mapping as well as in discovering sites of DNA/protein interaction (ChIP-chip, DamID), of DNA methylation (MeDIP-chip) and of sensitivity to DNase (DNase Chip) and array CGH.[1] In addition to detecting previously unidentified genes and regulatory sequences, improved quantification of transcription products is possible. Specific probes are present in millions of copies (as opposed to only several in traditional arrays) within an array unit called a feature, with anywhere from 10,000 to more than 6,000,000 different features per array.[2] Variable mapping resolutions are obtainable by adjusting the amount of sequence overlap between probes, or the amount of known base pairs between probe sequences, as well as probe length. For smaller genomes such as Arabidopsis, whole genomes can be examined.[3] Tiling arrays are a useful tool in genome-wide association studies.
Synthesis and manufacturers
The two main ways of synthesizing tiling arrays are photolithographic manufacturing and mechanical spotting or printing.
The first method involves in situ synthesis where probes, approximately 25bp, are built on the surface of the chip. These arrays can hold up to 6 million discrete features, each of which contains millions of copies of one probe.
The other way of synthesizing tiling array chips is via mechanically printing probes onto the chip. This is done by using automated machines with pins that place the previously synthesized probes onto the surface. Due to the size restriction of the pins, these chips can hold up to nearly 400,000 features.[4] Three manufacturers of tiling arrays are Affymetrix, NimbleGen and Agilent. Their products vary in probe length and spacing. ArrayExplorer.com is a free web-server to compare tiling arrays.
Applications and types
ChIP-chip
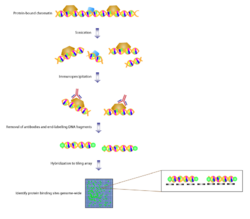
ChIP-chip is one of the most popular usages of tiling arrays. Chromatin immunoprecipitation allows binding sites of proteins to be identified. A genome-wide variation of this is known as ChIP-on-chip. Proteins that bind to chromatin are cross-linked in vivo, usually via fixation with formaldehyde. The chromatin is then fragmented and exposed to antibodies specific to the protein of interest. These complexes are then precipitated. The DNA is then isolated and purified. With traditional DNA microarrays, the immunoprecipitated DNA is hybridized to the chip, which contains probes that are designed to cover representative genome regions. Overlapping probes or probes in very close proximity can be used. This gives an unbiased analysis with high resolution. Besides these advantages, tiling arrays show high reproducibility and with overlapping probes spanning large segments of the genome, tiling arrays can interrogate protein binding sites, which harbor repeats. ChIP-chip experiments have been able to identify binding sites of transcription factors across the genome in yeast, drosophila and a few mammalian species.[5]
Transcriptome mapping
Another popular use of tiling arrays is in finding expressed genes. Traditional methods of gene prediction for annotation of genomic sequences have had problems when used to map the transcriptome, such as not producing an accurate structure of the genes and also missing transcripts entirely. The method of sequencing cDNA to find transcribed genes also runs into problems, such as failing to detect rare or very short RNA molecules, and so do not detect genes that are active only in response to signals or specific to a time frame. Tiling arrays can solve these issues. Due to the high resolution and sensitivity, even small and rare molecules can be detected. The overlapping nature of the probes also allows detection of non-polyadenylated RNA and can produce a more precise picture of gene structure.[6] Earlier studies on chromosome 21 and 22 showed the power of tiling arrays for identifying transcription units.[7][8][9] The authors used 25-mer probes that were 35bp apart, spanning the entire chromosomes. Labeled targets were made from polyadenylated RNA. They found many more transcripts than predicted and 90% were outside of annotated exons. Another study with Arabidopsis used high-density oligonucleotide arrays that cover the entire genome. More than 10 times more transcripts were found than predicted by ESTs[clarification needed] and other prediction tools.[3][10] Also found were novel transcripts in the centromeric regions where it was thought that no genes are actively expressed. Many noncoding and natural antisense RNA have been identified using tiling arrays.[9]
MeDIP-chip
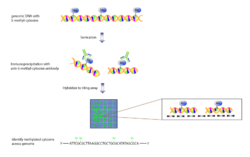
Methyl-DNA immunoprecipitation followed by tiling array allows DNA methylation mapping and measurement across the genome. DNA is methylated on cytosine in CG di-nucleotides in many places in the genome. This modification is one of the best-understood inherited epigenetic changes and is shown to affect gene expression. Mapping these sites can add to the knowledge of expressed genes and also epigenetic regulation on a genome-wide level. Tiling array studies have generated high-resolution methylation maps for the Arabidopsis genome to generate the first "methylome".
DNase-chip
DNase chip is an application of tiling arrays to identify hypersensitive sites, segments of open chromatin that are more readily cleaved by DNaseI. DNaseI cleaving produces larger fragments of around 1.2kb in size. These hypersensitive sites have been shown to accurately predict regulatory elements such as promoter regions, enhancers and silencers.[11] Historically, the method uses Southern blotting to find digested fragments. Tiling arrays have allowed researchers to apply the technique on a genome-wide scale.
Comparative genomic hybridization (CGH)
Array-based CGH is a technique often used in diagnostics to compare differences between types of DNA, such as normal cells vs. cancer cells. Two types of tiling arrays are commonly used for array CGH, whole genome and fine tiled. The whole genome approach would be useful in identifying copy number variations with high resolution. On the other hand, fine-tiled array CGH would produce ultrahigh resolution to find other abnormalities such as breakpoints.[12]
Procedure
Several different methods exist for tiling an array. One protocol for analyzing gene expression involves first isolating total RNA. This is then purified of rRNA molecules. The RNA is copied into double stranded DNA, which is subsequently amplified and in vitro transcribed to cRNA. The product is split into triplicates to produce dsDNA, which is then fragmented and labeled. Finally, the samples are hybridized to the tiling array chip. The signals from the chip are scanned and interpreted by computers.
Various software and algorithms are available for data analysis and vary in benefits depending on the manufacturer of the chip. For Affymetrix chips, the model-based analysis of tiling array (MAT) or hypergeometric analysis of tiling-arrays (HAT[13]) are effective peak-seeking algorithms. For NimbleGen chips, TAMAL is more suitable for locating binding sites. Alternative algorithms include MA2C and TileScope, which are less complicated to operate. The Joint binding deconvolution algorithm is commonly used for Agilent chips. If sequence analysis of binding site or annotation of the genome is required then programs like MEME, Gibbs Motif Sampler, Cis-regulatory element annotation system and Galaxy are used.[4]
Advantages and disadvantages
Tiling arrays provide an unbiased tool to investigate protein binding, gene expression and gene structure on a genome-wide scope. They allow a new level of insight in studying the transcriptome and methylome.
Drawbacks include the cost of tiling array kits. Although prices have fallen in the last several years, the price makes it impractical to use genome-wide tiling arrays for mammalian and other large genomes. Another issue is the "transcriptional noise" produced by its ultra-sensitive detection capability.[2] Furthermore, the approach provides no clearly defined start or stop to regions of interest identified by the array. Finally, arrays usually give only chromosome and position numbers, often necessitating sequencing as a separate step (although some modern arrays do give sequence information.[14])
References
- ↑ Yazaki, J; Gregory, BD; Ecker, JR (October 2007). "Mapping the genome landscape using tiling array technology.". Current Opinion in Plant Biology 10 (5): 534–42. doi:10.1016/j.pbi.2007.07.006. PMID 17703988.
- ↑ 2.0 2.1 Mockler, TC; Chan, S; Sundaresan, A; Chen, H; Jacobsen, SE; Ecker, JR (January 2005). "Applications of DNA tiling arrays for whole-genome analysis.". Genomics 85 (1): 1–15. doi:10.1016/j.ygeno.2004.10.005. PMID 15607417.
- ↑ 3.0 3.1 Yamada, K et al. (Oct 31, 2003). "Empirical analysis of transcriptional activity in the Arabidopsis genome.". Science 302 (5646): 842–6. doi:10.1126/science.1088305. PMID 14593172. Bibcode: 2003Sci...302..842Y.
- ↑ 4.0 4.1 Liu, XS (October 2007). "Getting started in tiling microarray analysis.". PLOS Computational Biology 3 (10): 1842–4. doi:10.1371/journal.pcbi.0030183. PMID 17967045. Bibcode: 2007PLSCB...3..183L.
- ↑ O'Geen, H; Squazzo, SL; Iyengar, S; Blahnik, K; Rinn, JL; Chang, HY; Green, R; Farnham, PJ (June 2007). "Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs.". PLOS Genetics 3 (6): e89. doi:10.1371/journal.pgen.0030089. PMID 17542650.
- ↑ Bertone, P; Gerstein, M; Snyder, M (2005). "Applications of DNA tiling arrays to experimental genome annotation and regulatory pathway discovery.". Chromosome Research 13 (3): 259–74. doi:10.1007/s10577-005-2165-0. PMID 15868420.
- ↑ Cawley, S; Bekiranov, S; Ng, HH; Kapranov, P; Sekinger, EA; Kampa, D; Piccolboni, A; Sementchenko, V et al. (Feb 20, 2004). "Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs.". Cell 116 (4): 499–509. doi:10.1016/S0092-8674(04)00127-8. PMID 14980218.
- ↑ Kapranov, P; Cawley, SE; Drenkow, J; Bekiranov, S; Strausberg, RL; Fodor, SP; Gingeras, TR (May 3, 2002). "Large-scale transcriptional activity in chromosomes 21 and 22.". Science 296 (5569): 916–9. doi:10.1126/science.1068597. PMID 11988577. Bibcode: 2002Sci...296..916K.
- ↑ 9.0 9.1 Kampa, D; Cheng, J; Kapranov, P; Yamanaka, M; Brubaker, S; Cawley, S; Drenkow, J; Piccolboni, A et al. (March 2004). "Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22.". Genome Research 14 (3): 331–42. doi:10.1101/gr.2094104. PMID 14993201.
- ↑ Stolc, V; Samanta, MP; Tongprasit, W; Sethi, H; Liang, S; Nelson, DC; Hegeman, A; Nelson, C et al. (Mar 22, 2005). "Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays.". Proceedings of the National Academy of Sciences of the United States of America 102 (12): 4453–8. doi:10.1073/pnas.0408203102. PMID 15755812. Bibcode: 2005PNAS..102.4453S.
- ↑ Crawford, Gregory E; Davis, Sean; Scacheri, Peter C; Renaud, Gabriel; Halawi, Mohamad J; Erdos, Michael R; Green, Roland; Meltzer, Paul S et al. (21 June 2006). "DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays". Nature Methods 3 (7): 503–9. doi:10.1038/nmeth888. PMID 16791207.
- ↑ Heidenblad, M; Lindgren, D; Jonson, T; Liedberg, F; Veerla, S; Chebil, G; Gudjonsson, S; Borg, A et al. (Jan 31, 2008). "Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors.". BMC Medical Genomics 1: 3. doi:10.1186/1755-8794-1-3. PMID 18237450.
- ↑ Taskesen, Erdogan; Beekman, Renee; de Ridder, Jeroen; Wouters, Bas J; Peeters, Justine K; Touw, Ivo P; Reinders, Marcel J.T; Delwel, Ruud (2010). "HAT: Hypergeometric Analysis of Tiling-arrays with application to promoter-GeneChip data". BMC Bioinformatics 11 (1): 275. doi:10.1186/1471-2105-11-275. PMID 20492700.
- ↑ Mockler, Todd C.; Ecker, Joseph R. (January 2005). "Applications of DNA tiling arrays for whole-genome analysis". Genomics 85 (1): 1–15. doi:10.1016/j.ygeno.2004.10.005. PMID 15607417.
 |