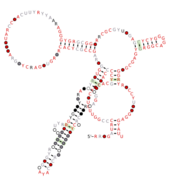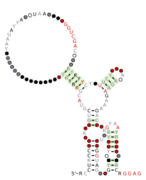Biology:LysM RNA motifs
lysM RNA motifs are conserved RNA structures that were discovered by bioinformatics.[1] Such bacterial motifs are defined by consistently being upstream of 'lysM' genes, which encode lysin protein domains, a conserved domain that participates in cell wall degradation. lysM motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes, although this hypothesis is not certain.
Three lysM RNA motifs have been found. lysM-Actino motif RNAs are found in Actinomycetales. lysM-Prevotella motif RNAs are found in the genus Prevotella. The lysM-TM7 RNA motif occurs only in the poorly understood phylum TM7.
A part of the lysM-Actino motif is likely the Shine-Dalgarno sequence of the downstream lysM gene. Thus, these RNAs might regulate the downstream gene translationally.
References
- ↑ "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Research 45 (18): 10811–10823. October 2017. doi:10.1093/nar/gkx699. PMID 28977401.
 |